ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3
- PMID: 23742014
- PMCID: PMC3823012
- DOI: 10.1111/tpj.12259
ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3
Abstract
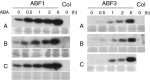
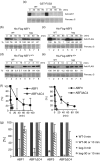
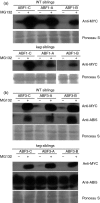
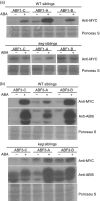
The ABA Binding Factor/ABA-Responsive Element Binding Proteins (ABF/AREB) subfamily of bZIP-type transcription factors are positive effectors of ABA responses. Here, we examine the proteolytic regulation of two members: Arabidopsis thaliana ABF1 and ABF3. Both transcription factors are unstable in seedlings, and their degradation is sensitive to proteasome inhibition. ABA treatment of seedlings leads to their rapid accumulation, the result of slowed proteolysis. Deletion of the conserved C-terminal region required for 14-3-3 interaction destabilizes the proteins. The degradation of ABF1 and ABF3 are slower in vivo in seedlings lacking the ubiquitin E3 ligase KEEP ON GOING (KEG), and in vitro in extracts from keg seedlings, implicating KEG in their degradation. ABF1 and ABF3 are ubiquitylation substrates of KEG in vitro, and in vitro pull-down assays document their direct interaction. In contrast to ABI5, another KEG substrate, the degradation of ABFs and proteolytic regulation of ABFs by ABA still occurs in keg seedlings, suggesting that additional E3s participate in ABF1 and ABF3 proteolysis. Loss of ABF1 or ABF3 in the keg background has a phenotypic effect similar to the loss of ABI5, and there is no additional rescue of the keg phenotype in abf1 abf3 abi5 keg seedlings. This result suggests that the abundance of other substrates is altered in keg seedlings, affecting growth. In conclusion, ABF1 and ABF3 abundance is affected by ABA and KEG, and the conserved C4 region serves as a stabilizing element.
Keywords: ABF; Arabidopsis thaliana; KEG; abscisic acid; proteolysis; ubiquitin; ubiquitin E3 ligase.
© 2013 The Authors The Plant Journal © 2013 John Wiley & Sons Ltd.
Figures





Similar articles
-
Factors that affect protein abundance of a positive regulator of abscisic acid signalling, the basic leucine zipper transcription factor ABRE-binding factor 2 (ABF2).Plant Direct. 2021 Jun 30;5(6):e00330. doi: 10.1002/pld3.330. eCollection 2021 Jun. Plant Direct. 2021. PMID: 34222769 Free PMC article.
-
Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation.Plant Cell. 2010 Aug;22(8):2630-41. doi: 10.1105/tpc.110.076075. Epub 2010 Aug 3. Plant Cell. 2010. PMID: 20682837 Free PMC article.
-
Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING.J Biol Chem. 2013 Jul 12;288(28):20267-79. doi: 10.1074/jbc.M113.465369. Epub 2013 May 29. J Biol Chem. 2013. PMID: 23720747 Free PMC article.
-
Structural Dynamics, Evolutionary Significance, and Functions of Really Interesting New Gene Proteins in Ubiquitination and Plant Stress: A Review.DNA Cell Biol. 2025 May;44(5):214-228. doi: 10.1089/dna.2025.0002. Epub 2025 Apr 10. DNA Cell Biol. 2025. PMID: 40208634 Review.
-
Ubiquitylation of ABA Receptors and Protein Phosphatase 2C Coreceptors to Modulate ABA Signaling and Stress Response.Int J Mol Sci. 2021 Jul 1;22(13):7103. doi: 10.3390/ijms22137103. Int J Mol Sci. 2021. PMID: 34281157 Free PMC article. Review.
Cited by
-
Comparative transcriptome analysis highlights the hormone effects on somatic embryogenesis in Catalpa bungei.Plant Reprod. 2019 Jun;32(2):141-151. doi: 10.1007/s00497-018-0349-y. Epub 2018 Nov 12. Plant Reprod. 2019. PMID: 30421145
-
OsDIRP1, a Putative RING E3 Ligase, Plays an Opposite Role in Drought and Cold Stress Responses as a Negative and Positive Factor, Respectively, in Rice (Oryza sativa L.).Front Plant Sci. 2018 Dec 5;9:1797. doi: 10.3389/fpls.2018.01797. eCollection 2018. Front Plant Sci. 2018. PMID: 30568669 Free PMC article.
-
Characterization and regulation mechanism analysis of ubiquitin-conjugating family genes in strawberry reveals a potential role in fruit ripening.BMC Plant Biol. 2022 Jan 19;22(1):39. doi: 10.1186/s12870-021-03421-8. BMC Plant Biol. 2022. PMID: 35045827 Free PMC article.
-
The Kinase Activity of Calcineurin B-like Interacting Protein Kinase 26 (CIPK26) Influences Its Own Stability and that of the ABA-regulated Ubiquitin Ligase, Keep on Going (KEG).Front Plant Sci. 2017 Apr 10;8:502. doi: 10.3389/fpls.2017.00502. eCollection 2017. Front Plant Sci. 2017. PMID: 28443108 Free PMC article.
-
Control of Amino Acid Homeostasis by a Ubiquitin Ligase-Coactivator Protein Complex.J Biol Chem. 2017 Mar 3;292(9):3827-3840. doi: 10.1074/jbc.M116.766469. Epub 2017 Jan 18. J Biol Chem. 2017. PMID: 28100770 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

