Engineering RNA-binding proteins for biology
- PMID: 23742071
- PMCID: PMC4052831
- DOI: 10.1111/febs.12375
Engineering RNA-binding proteins for biology
Abstract
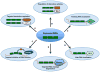
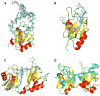
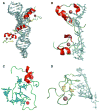
RNA-binding proteins play essential roles in the regulation of gene expression. Many have modular structures and combine relatively few common domains in various arrangements to recognize RNA sequences and/or structures. Recent progress in engineering the specificity of the PUF class RNA-binding proteins has shown that RNA-binding domains may be combined with various effector or functional domains to regulate the metabolism of targeted RNAs. Designer RNA-binding proteins with tailored sequence specificity will provide valuable tools for biochemical research as well as potential therapeutic applications. In this review, we discuss the suitability of various RNA-binding domains for engineering RNA-binding specificity, based on the structural basis for their recognition. We also compare various protein engineering and design methods applied to RNA-binding proteins, and discuss future applications of these proteins.
Keywords: RNA recognition motif; RNA-binding domains; RNA-binding proteins; computational design; in vitro evolution; phage display; protein engineering; protein-RNA interactions; yeast three-hybrid system; zinc finger.
© 2013 FEBS.
Figures


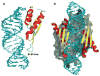


References
-
- Chen Y, Varani G. Protein families and RNA recognition. FEBS J. 2005;272:2088–2097. - PubMed
-
- Pavletich NP, Pabo CO. Zinc finger–DNA recognition: crystal structure of a Zif268–DNA complex at 2.1 Å. Science. 1991;252:809–817. - PubMed
-
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annu Rev Biochem. 2010;79:213–231. - PubMed
-
- Jamieson AC, Miller JC, Pabo CO. Drug discovery with engineered zinc-finger proteins. Nat Rev Drug Discov. 2003;2:361–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

