Unearthing the root of amino acid similarity
- PMID: 23743923
- PMCID: PMC6763418
- DOI: 10.1007/s00239-013-9565-0
Unearthing the root of amino acid similarity
Abstract
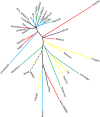
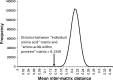
Similarities and differences between amino acids define the rates at which they substitute for one another within protein sequences and the patterns by which these sequences form protein structures. However, there exist many ways to measure similarity, whether one considers the molecular attributes of individual amino acids, the roles that they play within proteins, or some nuanced contribution of each. One popular approach to representing these relationships is to divide the 20 amino acids of the standard genetic code into groups, thereby forming a simplified amino acid alphabet. Here, we develop a method to compare or combine different simplified alphabets, and apply it to 34 simplified alphabets from the scientific literature. We use this method to show that while different suggestions vary and agree in non-intuitive ways, they combine to reveal a consensus view of amino acid similarity that is clearly rooted in physico-chemistry.
Figures




Similar articles
-
Evolution as a Guide to Designing xeno Amino Acid Alphabets.Int J Mol Sci. 2021 Mar 10;22(6):2787. doi: 10.3390/ijms22062787. Int J Mol Sci. 2021. PMID: 33801827 Free PMC article. Review.
-
Grouping of amino acids and recognition of protein structurally conserved regions by reduced alphabets of amino acids.Sci China C Life Sci. 2007 Jun;50(3):392-402. doi: 10.1007/s11427-007-0023-3. Sci China C Life Sci. 2007. PMID: 17609897
-
Reduced amino acid alphabets exhibit an improved sensitivity and selectivity in fold assignment.Bioinformatics. 2009 Jun 1;25(11):1356-62. doi: 10.1093/bioinformatics/btp164. Epub 2009 Apr 7. Bioinformatics. 2009. PMID: 19351620 Free PMC article.
-
Accuracy of sequence alignment and fold assessment using reduced amino acid alphabets.Proteins. 2006 Jun 1;63(4):986-95. doi: 10.1002/prot.20881. Proteins. 2006. PMID: 16506243
-
Knowledge-based potential functions in protein design.Curr Opin Struct Biol. 2002 Aug;12(4):447-52. doi: 10.1016/s0959-440x(02)00346-9. Curr Opin Struct Biol. 2002. PMID: 12163066 Review.
Cited by
-
Candida albicans' inorganic phosphate transport and evolutionary adaptation to phosphate scarcity.PLoS Genet. 2024 Aug 13;20(8):e1011156. doi: 10.1371/journal.pgen.1011156. eCollection 2024 Aug. PLoS Genet. 2024. PMID: 39137212 Free PMC article.
-
Adaptive Properties of the Genetically Encoded Amino Acid Alphabet Are Inherited from Its Subsets.Sci Rep. 2019 Aug 28;9(1):12468. doi: 10.1038/s41598-019-47574-x. Sci Rep. 2019. PMID: 31462646 Free PMC article.
-
Interpretable Machine Learning of Amino Acid Patterns in Proteins: A Statistical Ensemble Approach.J Chem Theory Comput. 2023 Sep 12;19(17):6011-6022. doi: 10.1021/acs.jctc.3c00383. Epub 2023 Aug 8. J Chem Theory Comput. 2023. PMID: 37552831 Free PMC article.
-
Experimental solutions to problems defining the origin of codon-directed protein synthesis.Biosystems. 2019 Sep;183:103979. doi: 10.1016/j.biosystems.2019.103979. Epub 2019 Jun 6. Biosystems. 2019. PMID: 31176803 Free PMC article. Review.
-
Evolution as a Guide to Designing xeno Amino Acid Alphabets.Int J Mol Sci. 2021 Mar 10;22(6):2787. doi: 10.3390/ijms22062787. Int J Mol Sci. 2021. PMID: 33801827 Free PMC article. Review.
References
-
- Andersen CAF, Brunak S. Representation of protein-sequence information by amino acid subalphabets. AI Magazine. 2004;25:97–104.
-
- Betts MJ, Russell RB. Bioinformatics for geneticists. New York: Wiley; 2003. Amino acid properties and consequences of substitutions.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

