Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli
- PMID: 23744946
- PMCID: PMC3777776
- DOI: 10.1126/science.1234012
Structural systems biology evaluation of metabolic thermotolerance in Escherichia coli
Abstract
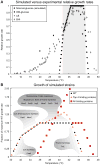
Genome-scale network reconstruction has enabled predictive modeling of metabolism for many systems. Traditionally, protein structural information has not been represented in such reconstructions. Expansion of a genome-scale model of Escherichia coli metabolism by including experimental and predicted protein structures enabled the analysis of protein thermostability in a network context. This analysis allowed the prediction of protein activities that limit network function at superoptimal temperatures and mechanistic interpretations of mutations found in strains adapted to heat. Predicted growth-limiting factors for thermotolerance were validated through nutrient supplementation experiments and defined metabolic sensitivities to heat stress, providing evidence that metabolic enzyme thermostability is rate-limiting at superoptimal temperatures. Inclusion of structural information expanded the content and predictive capability of genome-scale metabolic networks that enable structural systems biology of metabolism.
Figures



References
-
- Van Derlinden E, Bernaerts K, Van Impe JF. Dynamics of Escherichia coli at elevated temperatures: effect of temperature history and medium. J Appl Microbiol. 2008 Feb;104:438. - PubMed
-
- Fischer CR, Klein-Marcuschamer D, Stephanopoulos G. Selection and optimization of microbial hosts for biofuels production. Metab Eng. 2008 Nov;10:295. - PubMed
-
- Beltrao P, Kiel C, Serrano L. Structures in systems biology. Curr Opin Struct Biol. 2007 Jun;17:378. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

