Hydrogen tunneling links protein dynamics to enzyme catalysis
- PMID: 23746260
- PMCID: PMC4066974
- DOI: 10.1146/annurev-biochem-051710-133623
Hydrogen tunneling links protein dynamics to enzyme catalysis
Abstract
The relationship between protein dynamics and function is a subject of considerable contemporary interest. Although protein motions are frequently observed during ligand binding and release steps, the contribution of protein motions to the catalysis of bond making/breaking processes is more difficult to probe and verify. Here, we show how the quantum mechanical hydrogen tunneling associated with enzymatic C-H bond cleavage provides a unique window into the necessity of protein dynamics for achieving optimal catalysis. Experimental findings support a hierarchy of thermodynamically equilibrated motions that control the H-donor and -acceptor distance and active-site electrostatics, creating an ensemble of conformations suitable for H-tunneling. A possible extension of this view to methyl transfer and other catalyzed reactions is also presented. The impact of understanding these dynamics on the conceptual framework for enzyme activity, inhibitor/drug design, and biomimetic catalyst design is likely to be substantial.
Figures

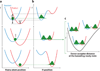
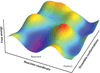
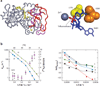



References
-
- Pauling L. Chemical achievement and hope for the future. Am. Sci. 1948;36:51–58. - PubMed
-
- Wolfenden R. Degrees of difficulty of water-consuming reactions in the absence of enzymes. Chem. Rev. 2006;106:3379–3396. - PubMed
-
- Hilvert D. Critical analysis of antibody catalysis. Annu. Rev. Biochem. 2000;69:751–793. - PubMed
-
- Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu. Rev. Biochem. 2006;75:519–541. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

