Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment
- PMID: 23746350
- PMCID: PMC3679449
- DOI: 10.1016/j.molcel.2013.05.011
Kinetochores coordinate pericentromeric cohesion and early DNA replication by Cdc7-Dbf4 kinase recruitment
Abstract
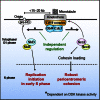
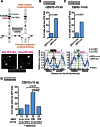
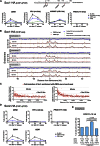
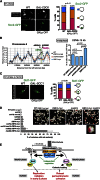
Centromeres play several important roles in ensuring proper chromosome segregation. Not only do they promote kinetochore assembly for microtubule attachment, but they also support robust sister chromatid cohesion at pericentromeres and facilitate replication of centromeric DNA early in S phase. However, it is still elusive how centromeres orchestrate all these functions at the same site. Here, we show that the budding yeast Dbf4-dependent kinase (DDK) accumulates at kinetochores in telophase, facilitated by the Ctf19 kinetochore complex. This promptly recruits Sld3-Sld7 replication initiator proteins to pericentromeric replication origins so that they initiate replication early in S phase. Furthermore, DDK at kinetochores independently recruits the Scc2-Scc4 cohesin loader to centromeres in G1 phase. This enhances cohesin loading and facilitates robust pericentromeric cohesion in S phase. Thus, we have found the central mechanism by which kinetochores orchestrate early S phase DNA replication and robust sister chromatid cohesion at microtubule attachment sites.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures




References
-
- Araki H. Initiation of chromosomal DNA replication in eukaryotic cells; contribution of yeast genetics to the elucidation. Genes Genet. Syst. 2011;86:141–149. - PubMed
-
- Bailis J.M., Bernard P., Antonelli R., Allshire R.C., Forsburg S.L. Hsk1-Dfp1 is required for heterochromatin-mediated cohesion at centromeres. Nat. Cell Biol. 2003;5:1111–1116. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

