Blood RNA profiling in a large cohort of multiple sclerosis patients and healthy controls
- PMID: 23748426
- PMCID: PMC3781642
- DOI: 10.1093/hmg/ddt267
Blood RNA profiling in a large cohort of multiple sclerosis patients and healthy controls
Abstract
Multiple sclerosis (MS) is the most common autoimmune disease of the central nervous system (CNS). It is characterized by the infiltration of autoreactive immune cells into the CNS, which target the myelin sheath, leading to the loss of neuronal function. Although it is accepted that MS is a multifactorial disorder with both genetic and environmental factors influencing its development and course, the molecular pathogenesis of MS has not yet been fully elucidated. Here, we studied the longitudinal gene expression profiles of whole-blood RNA from a cohort of 195 MS patients and 66 healthy controls. We analyzed these transcriptomes at both the individual transcript and the biological pathway level. We found 62 transcripts to be significantly up-regulated in MS patients; the expression of 11 of these genes was counter-regulated by interferon treatment, suggesting partial restoration of a 'healthy' gene expression profile. Global pathway analyses linked the proteasome and Wnt signaling to MS disease processes. Since genotypes from a subset of individuals were available, we were able to identify expression quantitative trait loci (eQTL), a number of which involved two genes of the MS gene signature. However, all these eQTL were also present in healthy controls. This study highlights the challenge posed by analyzing transcripts from whole blood and how these can be mitigated by using large, well-characterized cohorts of patients with longitudinal follow-up and multi-modality measurements.
Figures

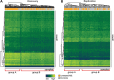

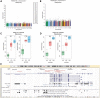
References
-
- Rosati G. The prevalence of multiple sclerosis in the world: an update. Neurol. Sci. 2001;22:117–139. - PubMed
-
- Sospedra M., Martin R. Immunology of multiple sclerosis. Annu. Rev. Immunol. 2005;23:683–747. - PubMed
-
- Oksenberg J.R., Baranzini S.E. Multiple sclerosis genetics—is the glass half full, or half empty? Nat. Rev. Neurol. 2010;6:429–437. - PubMed
-
- Haines J.L., Ter-Minassian M., Bazyk A., Gusella J.F., Kim D.J., Terwedow H., Pericak-Vance M.A., Rimmler J.B., Haynes C.S., Roses A.D., et al. A complete genomic screen for multiple sclerosis underscores a role for the major histocompatability complex. The Multiple Sclerosis Genetics Group. Nat. Genet. 1996;13:469–471. - PubMed
-
- Nagasaka Y., Dillner K., Ebise H., Teramoto R., Nakagawa H., Lilius L., Axelman K., Forsell C., Ito A., Winblad B., et al. A unique gene expression signature discriminates familial Alzheimer's disease mutation carriers from their wild-type siblings. Proc. Natl Acad. Sci. USA. 2005;102:14854–14859. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

