The blockade of the neurotransmitter release apparatus by botulinum neurotoxins
- PMID: 23749048
- PMCID: PMC11113401
- DOI: 10.1007/s00018-013-1380-7
The blockade of the neurotransmitter release apparatus by botulinum neurotoxins
Abstract
The high toxicity of the seven serotypes of botulinum neurotoxins (BoNT/A to G), together with their specificity and reversibility, includes them in the list A of potential bioterrorism weapons and, at the same time, among the therapeutics of choice for a variety of human syndromes. They invade nerve terminals and cleave specifically the three proteins which form the heterotrimeric SNAP REceptors (SNARE) complex that mediates neurotransmitter release. The BoNT-induced cleavage of the SNARE proteins explains by itself the paralysing activity of the BoNTs because the truncated proteins cannot form the SNARE complex. However, in the case of BoNT/A, the most widely used toxin in therapy, additional factors come into play as it only removes a few residues from the synaptosomal associate protein of 25 kDa C-terminus and this results in a long duration of action. To explain these facts and other experimental data, we present here a model for the assembly of the neuroexocytosis apparatus in which Synaptotagmin and Complexin first assist the zippering of the SNARE complex, and then stabilize and clamp an octameric radial assembly of the SNARE complexes.
Figures
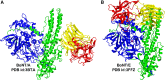


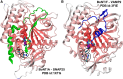


References
-
- Hill KK, Smith TJ. Genetic diversity within Clostridium botulinum serotypes, botulinum neurotoxin gene clusters and toxin subtypes. Curr Top Microbiol Immunol. 2013;364:1–20. - PubMed
-
- Johnson EA, Montecucco C. Botulism. Handb Clin Neurol. 2008;91:333–368. - PubMed
-
- Caleo M, Schiavo G. Central effects of tetanus and botulinum neurotoxins. Toxicon. 2009;54:593–599. - PubMed
-
- Schiavo G, Matteoli M, Montecucco C. Neurotoxins affecting neuroexocytosis. Physiol Rev. 2000;80:717–766. - PubMed
-
- Takamori S, Holt M, Stenius K, Lemke EA, Gronborg M, Riedel D, Urlaub H, Schenck S, Brugger B, Ringler P, Muller SA, Rammner B, Grater F, Hub JS, De Groot BL, Mieskes G, Moriyama Y, Klingauf J, Grubmuller H, Heuser J, Wieland F, Jahn R. Molecular anatomy of a trafficking organelle. Cell. 2006;127:831–846. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

