PathAct: a novel method for pathway analysis using gene expression profiles
- PMID: 23750088
- PMCID: PMC3670121
- DOI: 10.6026/97320630009394
PathAct: a novel method for pathway analysis using gene expression profiles
Abstract
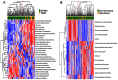
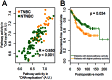
We developed PathAct, a novel method for pathway analysis to investigate the biological and clinical implications of the gene expression profiles. The advantage of PathAct in comparison with the conventional pathway analysis methods is that it can estimate pathway activity levels for individual patient quantitatively in the form of a pathway-by-sample matrix. This matrix can be used for further analysis such as hierarchical clustering and other analysis methods. To evaluate the feasibility of PathAct, comparison with frequently used gene-enrichment analysis methods was conducted using two public microarray datasets. The dataset #1 was that of breast cancer patients, and we investigated pathways associated with triple-negative breast cancer by PathAct, compared with those obtained by gene set enrichment analysis (GSEA). The dataset #2 was another breast cancer dataset with disease-free survival (DFS) of each patient. Contribution by each pathway to prognosis was investigated by our method as well as the Database for Annotation, Visualization and Integrated Discovery (DAVID) analysis. In the dataset #1, four out of the six pathways that satisfied p < 0.05 and FDR < 0.30 by GSEA were also included in those obtained by the PathAct method. For the dataset #2, two pathways ("Cell Cycle" and "DNA replication") out of four pathways by PathAct were commonly identified by DAVID analysis. Thus, we confirmed a good degree of agreement among PathAct and conventional methods. Moreover, several applications of further statistical analyses such as hierarchical cluster analysis by pathway activity, correlation analysis and survival analysis between pathways were conducted.
Figures
References
-
- Nishimura D, et al. Biotech Softw Internet Rep. 2001;2:117.
-
- Dahlquist KD, et al. Nat Genet. 2002;31:19. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
