Dynamic microtubules produce an asymmetric E-cadherin-Bazooka complex to maintain segment boundaries
- PMID: 23751496
- PMCID: PMC3678168
- DOI: 10.1083/jcb.201211159
Dynamic microtubules produce an asymmetric E-cadherin-Bazooka complex to maintain segment boundaries
Abstract
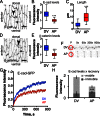
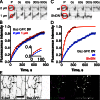
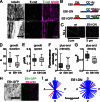
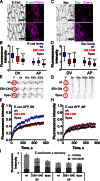
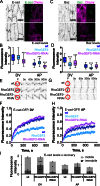
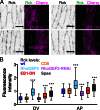
Distributing junctional components around the cell periphery is key for epithelial tissue morphogenesis and homeostasis. We discovered that positioning of dynamic microtubules controls the asymmetric accumulation of E-cadherin. Microtubules are oriented preferentially along the dorso-ventral axis in Drosophila melanogaster embryonic epidermal cells, and thus more frequently contact E-cadherin at dorso-ventral cell-cell borders. This inhibits RhoGEF2, reducing membrane recruitment of Rho-kinase, and increasing a specific E-cadherin pool that is mobile when assayed by fluorescence recovery after photobleaching. This mobile E-cadherin is complexed with Bazooka/Par-3, which in turn is required for normal levels of mobile E-cadherin. Mobile E-cadherin-Bazooka prevents formation of multicellular rosette structures and cell motility across the segment border in Drosophila embryos. Altogether, the combined action of dynamic microtubules and Rho signaling determines the level and asymmetric distribution of a mobile E-cadherin-Bazooka complex, which regulates cell behavior during the generation of a patterned epithelium.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

