Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics
- PMID: 23751777
- PMCID: PMC3919211
- DOI: 10.1053/j.gastro.2013.05.050
Beyond gene discovery in inflammatory bowel disease: the emerging role of epigenetics
Abstract
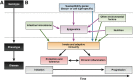
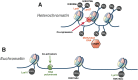
In the past decade, there have been fundamental advances in our understanding of genetic factors that contribute to the inflammatory bowel diseases (IBDs) Crohn's disease and ulcerative colitis. The latest international collaborative studies have brought the number of IBD susceptibility gene loci to 163. However, genetic factors account for only a portion of overall disease variance, indicating a need to better explore gene-environment interactions in the development of IBD. Epigenetic factors can mediate interactions between the environment and the genome; their study could provide new insight into the pathogenesis of IBD. We review recent progress in identification of genetic factors associated with IBD and discuss epigenetic mechanisms that could affect development and progression of IBD.
Keywords: CD; CpG; Crohn’s Disease; Crohn’s disease; DNA Methylation; DNA methyltransferase; DNMT; EWAS; Epigenetics; GWAS; HAT; HDAC; HDACi; IBD; IL; NF-κB; PBMC; SNP; T-helper; TNF; Th; UC; Ulcerative Colitis; cytosine-guanine dinucleotides; epigenome-wide methylation association studies; genome-wide association studies; histone acetyl transferase; histone deacetylase; histone deacetylatase inhibitors; inflammatory bowel disease; interleukin; mRNA; messenger RNA; miR; microRNA; nuclear factor κB; peripheral blood mononuclear cell; single nucleotide polymorphism; tumor necrosis factor; ulcerative colitis.
Copyright © 2013 AGA Institute. Published by Elsevier Inc. All rights reserved.
Figures


References
-
- Molodecky N.A., Soon I.S., Rabi D.M. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology. 2012;142:46–54.e42. quiz e30. - PubMed
-
- Henderson P., Hansen R., Cameron F.L. Rising incidence of pediatric inflammatory bowel disease in Scotland. Inflamm Bowel Dis. 2012;18:999–1005. - PubMed
-
- Benchimol E.I., Guttmann A., Griffiths A.M. Increasing incidence of paediatric inflammatory bowel disease in Ontario, Canada: evidence from health administrative data. Gut. 2009;58:1490–1497. - PubMed
-
- Ng S.C., Bernstein C.N., Vatn M.H. Geographical variability and environmental risk factors in inflammatory bowel disease. Gut. 2013;62:630–649. - PubMed
-
- Gunesh S., Thomas G.A., Williams G.T. The incidence of Crohn’s disease in Cardiff over the last 75 years: an update for 1996-2005. Aliment Pharmacol Ther. 2008;27:211–219. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

