Posttranscriptional regulation of PER1 underlies the oncogenic function of IREα
- PMID: 23752693
- PMCID: PMC3915716
- DOI: 10.1158/0008-5472.CAN-12-3989
Posttranscriptional regulation of PER1 underlies the oncogenic function of IREα
Abstract
Growing evidence supports a role for the unfolded protein response (UPR) in carcinogenesis; however, the precise molecular mechanisms underlying this phenomenon remain elusive. Herein, we identified the circadian clock PER1 mRNA as a novel substrate of the endoribonuclease activity of the UPR sensor IRE1α. Analysis of the mechanism shows that IRE1α endoribonuclease activity decreased PER1 mRNA in tumor cells without affecting PER1 gene transcription. Inhibition of IRE1α signaling using either siRNA-mediated silencing or a dominant-negative strategy prevented PER1 mRNA decay, reduced tumorigenesis, and increased survival, features that were reversed upon PER1 silencing. Clinically, patients showing reduced survival have lower levels of PER1 mRNA expression and increased splicing of XBP1, a known IRE-α substrate, thereby pointing toward an increased IRE1α activity in these patients. Hence, we describe a novel mechanism connecting the UPR and circadian clock components in tumor cells, thereby highlighting the importance of this interplay in tumor development.
©2013 AACR.
Conflict of interest statement
J.N. Sarkaria has a commercial research grant from Genentech, Basilea, Sanofi, and Merck. E. Chevet has a commercial research grant from Servier. No potential conflicts of interest were disclosed by the other authors.
Figures



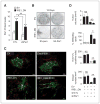

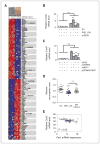
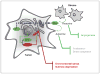
References
-
- Moenner M, Pluquet O, Bouchecareilh M, Chevet E. Integrated endoplasmic reticulum stress responses in cancer. Cancer Res. 2007;67:10631–4. - PubMed
-
- Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–89. - PubMed
-
- Bifulco G, Miele C, Di Jeso B, Beguinot F, Nappi C, Di Carlo C, et al. Endoplasmic reticulum stress is activated in endometrial adenocarcinoma. Gynecol Oncol. 2012;125:220–5. - PubMed
-
- Davies MP, Barraclough DL, Stewart C, Joyce KA, Eccles RM, Barraclough R, et al. Expression and splicing of the unfolded protein response gene XBP-1 are significantly associated with clinical outcome of endocrine-treated breast cancer. Int J Cancer. 2008;123:85–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

