Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum
- PMID: 23754396
- PMCID: PMC3696752
- DOI: 10.1073/pnas.1219809110
Candidate phylum TM6 genome recovered from a hospital sink biofilm provides genomic insights into this uncultivated phylum
Abstract
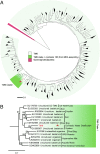
The "dark matter of life" describes microbes and even entire divisions of bacterial phyla that have evaded cultivation and have yet to be sequenced. We present a genome from the globally distributed but elusive candidate phylum TM6 and uncover its metabolic potential. TM6 was detected in a biofilm from a sink drain within a hospital restroom by analyzing cells using a highly automated single-cell genomics platform. We developed an approach for increasing throughput and effectively improving the likelihood of sampling rare events based on forming small random pools of single-flow-sorted cells, amplifying their DNA by multiple displacement amplification and sequencing all cells in the pool, creating a "mini-metagenome." A recently developed single-cell assembler, SPAdes, in combination with contig binning methods, allowed the reconstruction of genomes from these mini-metagenomes. A total of 1.07 Mb was recovered in seven contigs for this member of TM6 (JCVI TM6SC1), estimated to represent 90% of its genome. High nucleotide identity between a total of three TM6 genome drafts generated from pools that were independently captured, amplified, and assembled provided strong confirmation of a correct genomic sequence. TM6 is likely a Gram-negative organism and possibly a symbiont of an unknown host (nonfree living) in part based on its small genome, low-GC content, and lack of biosynthesis pathways for most amino acids and vitamins. Phylogenomic analysis of conserved single-copy genes confirms that TM6SC1 is a deeply branching phylum.
Keywords: MDA; genome assembly; metagenomics; single-cell genomics; symbiotic bacteria.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Lasken RS. Genomic sequencing of uncultured microorganisms from single cells. Nat Rev Microbiol. 2012;10(9):631–640. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

