Under-folded proteins: Conformational ensembles and their roles in protein folding, function, and pathogenesis
- PMID: 23754493
- PMCID: PMC7161862
- DOI: 10.1002/bip.22298
Under-folded proteins: Conformational ensembles and their roles in protein folding, function, and pathogenesis
Abstract
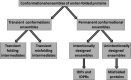
For decades, protein function was intimately linked to the presence of a unique, aperiodic crystal-like structure in a functional protein. The two only places for conformational ensembles of under-folded (or partially folded) protein forms in this picture were either the end points of the protein denaturation processes or transiently populated folding intermediates. Recent years witnessed dramatic change in this perception and conformational ensembles, which the under-folded proteins are, have moved from the shadow. Accumulated to date data suggest that a protein can exist in at least three global forms-functional and folded, functional and intrinsically disordered (nonfolded), and nonfunctional and misfolded/aggregated. Under-folded protein states are crucial for each of these forms, serving as important folding intermediates of ordered proteins, or as functional states of intrinsically disordered proteins (IDPs) and IDP regions (IDPRs), or as pathology triggers of misfolded proteins. Based on these observations, conformational ensembles of under-folded proteins can be classified as transient (folding and misfolding intermediates) and permanent (IDPs and stable misfolded proteins). Permanently under-folded proteins can further be split into intentionally designed (IDPs and IDPRs) and unintentionally designed (misfolded proteins). Although intrinsic flexibility, dynamics, and pliability are crucial for all under-folded proteins, the different categories of under-foldedness are differently encoded in protein amino acid sequences.
Keywords: conformational ensemble; intrinsically disordered protein; protein folding; protein misfolding.
Copyright © 2013 Wiley Periodicals, Inc.
Figures



Similar articles
-
Proteins without unique 3D structures: biotechnological applications of intrinsically unstable/disordered proteins.Biotechnol J. 2015 Mar;10(3):356-66. doi: 10.1002/biot.201400374. Epub 2014 Oct 6. Biotechnol J. 2015. PMID: 25287424 Review.
-
Intrinsically disordered proteins in crowded milieu: when chaos prevails within the cellular gumbo.Cell Mol Life Sci. 2018 Nov;75(21):3907-3929. doi: 10.1007/s00018-018-2894-9. Epub 2018 Jul 31. Cell Mol Life Sci. 2018. PMID: 30066087 Free PMC article. Review.
-
Conformational propensities of intrinsically disordered proteins influence the mechanism of binding and folding.Proc Natl Acad Sci U S A. 2015 Aug 4;112(31):9614-9. doi: 10.1073/pnas.1512799112. Epub 2015 Jul 20. Proc Natl Acad Sci U S A. 2015. PMID: 26195786 Free PMC article.
-
Functions of short lifetime biological structures at large: the case of intrinsically disordered proteins.Brief Funct Genomics. 2020 Jan 22;19(1):60-68. doi: 10.1093/bfgp/ely023. Brief Funct Genomics. 2020. PMID: 29982297 Review.
-
Order, Disorder, and Everything in Between.Molecules. 2016 Aug 19;21(8):1090. doi: 10.3390/molecules21081090. Molecules. 2016. PMID: 27548131 Free PMC article. Review.
Cited by
-
Molecular Serum Albumin Unmask Nanobio Properties of Molecular Graphenes in Shungite Carbon Nanoparticles.Int J Mol Sci. 2024 Feb 20;25(5):2465. doi: 10.3390/ijms25052465. Int J Mol Sci. 2024. PMID: 38473711 Free PMC article.
-
Ordered disorder of the astrocytic dystrophin-associated protein complex in the norm and pathology.PLoS One. 2013 Aug 27;8(8):e73476. doi: 10.1371/journal.pone.0073476. eCollection 2013. PLoS One. 2013. PMID: 24014171 Free PMC article.
-
Are Charge-State Distributions a Reliable Tool Describing Molecular Ensembles of Intrinsically Disordered Proteins by Native MS?J Am Soc Mass Spectrom. 2017 Jan;28(1):21-28. doi: 10.1007/s13361-016-1490-1. Epub 2016 Oct 11. J Am Soc Mass Spectrom. 2017. PMID: 27730522 Review.
-
Hbr1 Activates and Represses Hyphal Growth in Candida albicans and Regulates Fungal Morphogenesis under Embedded Conditions.PLoS One. 2015 Jun 3;10(6):e0126919. doi: 10.1371/journal.pone.0126919. eCollection 2015. PLoS One. 2015. PMID: 26039220 Free PMC article.
-
The protein disorder cycle.Biophys Rev. 2021 Nov 3;13(6):1155-1162. doi: 10.1007/s12551-021-00853-2. eCollection 2021 Dec. Biophys Rev. 2021. PMID: 35059033 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

