ATM mediates pRB function to control DNMT1 protein stability and DNA methylation
- PMID: 23754744
- PMCID: PMC3753918
- DOI: 10.1128/MCB.01597-12
ATM mediates pRB function to control DNMT1 protein stability and DNA methylation
Abstract
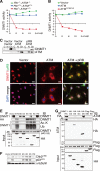
The retinoblastoma tumor suppressor gene (RB) product has been implicated in epigenetic control of gene expression owing to its ability to physically bind to many chromatin modifiers. However, the biological and clinical significance of this activity was not well elucidated. To address this, we performed genetic and epigenetic analyses in an Rb-deficient mouse thyroid C cell tumor model. Here we report that the genetic interaction of Rb and ATM regulates DNMT1 protein stability and hence controls the DNA methylation status in the promoters of at least the Ink4a, Shc2, FoxO6, and Noggin genes. Furthermore, we demonstrate that inactivation of pRB promotes Tip60 (acetyltransferase)-dependent ATM activation; allows activated ATM to physically bind to DNMT1, forming a complex with Tip60 and UHRF1 (E3 ligase); and consequently accelerates DNMT1 ubiquitination driven by Tip60-dependent acetylation. Our results indicate that inactivation of the pRB pathway in coordination with aberration in the DNA damage response deregulates DNMT1 stability, leading to an abnormal DNA methylation pattern and malignant progression.
Figures






References
-
- Robertson KD, Ait-Si-Ali S, Yokochi T, Wade PA, Jones PL, Wolffe AP. 2000. DNMT1 forms a complex with Rb, E2F1 and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 25:338–342 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
