A network of transcription factors operates during early tooth morphogenesis
- PMID: 23754753
- PMCID: PMC3753897
- DOI: 10.1128/MCB.00524-13
A network of transcription factors operates during early tooth morphogenesis
Abstract
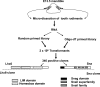
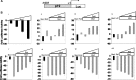
Improving the knowledge of disease-causing genes is a unique challenge in human health. Although it is known that genes causing similar diseases tend to lie close to one another in a network of protein-protein or functional interactions, the identification of these protein-protein networks is difficult to unravel. Here, we show that Msx1, Snail, Lhx6, Lhx8, Sp3, and Lef1 interact in vitro and in vivo, revealing the existence of a novel context-specific protein network. These proteins are all expressed in the neural crest-derived dental mesenchyme and cause tooth agenesis disorder when mutated in mouse and/or human. We also identified an in vivo direct target for Msx1 function, the cyclin D-dependent kinase (CDK) inhibitor p19(ink4d), whose transcription is differentially modulated by the protein network. Considering the important role of p19(ink4d) as a cell cycle regulator, these results provide evidence for the first time of the unique plasticity of the Msx1-dependent network of proteins in conferring differential transcriptional output and in controlling the cell cycle through the regulation of a cyclin D-dependent kinase inhibitor. Collectively, these data reveal a novel protein network operating in the neural crest-derived dental mesenchyme that is relevant for many other areas of developmental and evolutionary biology.
Figures








References
-
- Thesleff I, Vaahtokari A, Partanen AM. 1995. Regulation of organogenesis. Common molecular mechanisms regulating the development of teeth and other organs. Int. J. Dev. Biol. 39:35–50 - PubMed
-
- Bei M, Peters H, Maas RL. 2002. The role of PAX and MSX genes in craniofacial development, p 101–112 In Lin KY, Ogle RC, Jane JA. (ed), Craniofacial surgery: science & surgical technique. W. B. Saunders Company, Philadelphia, PA
-
- Hayashi S, Scott MP. 1990. What determines the specificity of action of Drosophila homeodomain proteins? Cell 63:883–894 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
