Impact of natural genetic variation on gene expression dynamics
- PMID: 23754949
- PMCID: PMC3674999
- DOI: 10.1371/journal.pgen.1003514
Impact of natural genetic variation on gene expression dynamics
Abstract
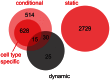
DNA sequence variation causes changes in gene expression, which in turn has profound effects on cellular states. These variations affect tissue development and may ultimately lead to pathological phenotypes. A genetic locus containing a sequence variation that affects gene expression is called an "expression quantitative trait locus" (eQTL). Whereas the impact of cellular context on expression levels in general is well established, a lot less is known about the cell-state specificity of eQTL. Previous studies differed with respect to how "dynamic eQTL" were defined. Here, we propose a unified framework distinguishing static, conditional and dynamic eQTL and suggest strategies for mapping these eQTL classes. Further, we introduce a new approach to simultaneously infer eQTL from different cell types. By using murine mRNA expression data from four stages of hematopoiesis and 14 related cellular traits, we demonstrate that static, conditional and dynamic eQTL, although derived from the same expression data, represent functionally distinct types of eQTL. While static eQTL affect generic cellular processes, non-static eQTL are more often involved in hematopoiesis and immune response. Our analysis revealed substantial effects of individual genetic variation on cell type-specific expression regulation. Among a total number of 3,941 eQTL we detected 2,729 static eQTL, 1,187 eQTL were conditionally active in one or several cell types, and 70 eQTL affected expression changes during cell type transitions. We also found evidence for feedback control mechanisms reverting the effect of an eQTL specifically in certain cell types. Loci correlated with hematological traits were enriched for conditional eQTL, thus, demonstrating the importance of conditional eQTL for understanding molecular mechanisms underlying physiological trait variation. The classification proposed here has the potential to streamline and unify future analysis of conditional and dynamic eQTL as well as many other kinds of QTL data.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
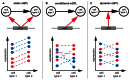

 . Similarly, for each condition the genotype matrix is subset to all samples for which there are expression measurements in this cell type. The resulting two submatrices
. Similarly, for each condition the genotype matrix is subset to all samples for which there are expression measurements in this cell type. The resulting two submatrices  and
and  are concatenated into one genotype matrix. In order to discriminate static and conditional eQTL, two additional predictors indicating the cell type from which a sample was derived, are added to the predictor matrix. The combined genotype and cell type indicator matrix is used to find the model which best predicts gene expression simultaneously in all conditions.
are concatenated into one genotype matrix. In order to discriminate static and conditional eQTL, two additional predictors indicating the cell type from which a sample was derived, are added to the predictor matrix. The combined genotype and cell type indicator matrix is used to find the model which best predicts gene expression simultaneously in all conditions.
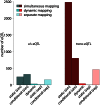
 shown separately for cis-eQTL (left) and trans-eQTL (right). Static, conditional and dynamic eQTL are distinguished (see labels at the bottom). Further, the figure discriminates simultaneous and separate eQTL mappings, which represent alternative ways for distinguishing static and conditional eQTL. Simultaneous mapping increases the statistical power leading to substantially more eQTL significant at the same level (
shown separately for cis-eQTL (left) and trans-eQTL (right). Static, conditional and dynamic eQTL are distinguished (see labels at the bottom). Further, the figure discriminates simultaneous and separate eQTL mappings, which represent alternative ways for distinguishing static and conditional eQTL. Simultaneous mapping increases the statistical power leading to substantially more eQTL significant at the same level ( ). Even though both, cis- and trans-eQTL are increased when performing simultaneous mapping, trans-eQTL benefit more from the increase in power. See main text for exact definitions of the various eQTL types.
). Even though both, cis- and trans-eQTL are increased when performing simultaneous mapping, trans-eQTL benefit more from the increase in power. See main text for exact definitions of the various eQTL types.

 ) are shown in gray, cell type-specific eQTL (
) are shown in gray, cell type-specific eQTL ( in exactly one cell type) are shown in the color scheme of Figure 3. Red triangles indicate two cell type-specific eQTL-rich regions (eQRR).
in exactly one cell type) are shown in the color scheme of Figure 3. Red triangles indicate two cell type-specific eQTL-rich regions (eQRR).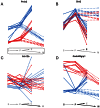
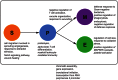
 ) among eQTL loci are shown in italic, GO categories enriched among eQTL targets in regular font. See Tables S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12 for a list of the top significant GO terms of each mapping.
) among eQTL loci are shown in italic, GO categories enriched among eQTL targets in regular font. See Tables S1, S2, S3, S4, S5, S6, S7, S8, S9, S10, S11, S12 for a list of the top significant GO terms of each mapping.References
-
- Dermitzakis ET (2008) From gene expression to disease risk. Nature Genetics 40: 492–493. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

