Inferring demographic history from a spectrum of shared haplotype lengths
- PMID: 23754952
- PMCID: PMC3675002
- DOI: 10.1371/journal.pgen.1003521
Inferring demographic history from a spectrum of shared haplotype lengths
Abstract
There has been much recent excitement about the use of genetics to elucidate ancestral history and demography. Whole genome data from humans and other species are revealing complex stories of divergence and admixture that were left undiscovered by previous smaller data sets. A central challenge is to estimate the timing of past admixture and divergence events, for example the time at which Neanderthals exchanged genetic material with humans and the time at which modern humans left Africa. Here, we present a method for using sequence data to jointly estimate the timing and magnitude of past admixture events, along with population divergence times and changes in effective population size. We infer demography from a collection of pairwise sequence alignments by summarizing their length distribution of tracts of identity by state (IBS) and maximizing an analytic composite likelihood derived from a Markovian coalescent approximation. Recent gene flow between populations leaves behind long tracts of identity by descent (IBD), and these tracts give our method power by influencing the distribution of shared IBS tracts. In simulated data, we accurately infer the timing and strength of admixture events, population size changes, and divergence times over a variety of ancient and recent time scales. Using the same technique, we analyze deeply sequenced trio parents from the 1000 Genomes project. The data show evidence of extensive gene flow between Africa and Europe after the time of divergence as well as substructure and gene flow among ancestral hominids. In particular, we infer that recent African-European gene flow and ancient ghost admixture into Europe are both necessary to explain the spectrum of IBS sharing in the trios, rejecting simpler models that contain less population structure.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
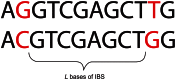
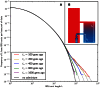
 base pairs of sequence alignment simulated with Hudson's MS . The IBS tracts are shared between two populations of constant size 10,000 that diverged 2,000 generations ago, with one haplotype sampled from each population. 5% of the genetic material from one population is the product of a recent admixture pulse from the other population. Figure 2B illustrates the history being simulated. When the admixture occurred less than 1,000 generations ago, it noticeably increases the abundance of long IBS tracts. The gray lines in 2A are theoretical tract abundance predictions, and fit the simulated data extremely well. To smooth out noise in the simulated data, abundances are averaged over intervals with exponentially spaced endpoints
base pairs of sequence alignment simulated with Hudson's MS . The IBS tracts are shared between two populations of constant size 10,000 that diverged 2,000 generations ago, with one haplotype sampled from each population. 5% of the genetic material from one population is the product of a recent admixture pulse from the other population. Figure 2B illustrates the history being simulated. When the admixture occurred less than 1,000 generations ago, it noticeably increases the abundance of long IBS tracts. The gray lines in 2A are theoretical tract abundance predictions, and fit the simulated data extremely well. To smooth out noise in the simulated data, abundances are averaged over intervals with exponentially spaced endpoints  .
.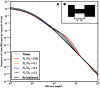
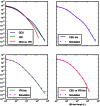
 base pairs of pairwise alignment. Both sequences are derived from the population depicted in Figure 3B that underwent a bottleneck from size
base pairs of pairwise alignment. Both sequences are derived from the population depicted in Figure 3B that underwent a bottleneck from size  to size
to size  , the duration of the bottleneck being
, the duration of the bottleneck being  generations. 1,000 generations ago, the population recovered to size 10,000. These bottlenecks leave similar frequencies of very long and very short IBS tracts because they have identical ratios of strength to duration, but they leave different signature increases compared to the no-bottleneck history in the abundance of
generations. 1,000 generations ago, the population recovered to size 10,000. These bottlenecks leave similar frequencies of very long and very short IBS tracts because they have identical ratios of strength to duration, but they leave different signature increases compared to the no-bottleneck history in the abundance of  –
– -base IBS tracts. In grey are the expected IBS tract spectra that we predict analytically for each simulated history.
-base IBS tracts. In grey are the expected IBS tract spectra that we predict analytically for each simulated history.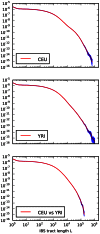
 -base IBS tracts observed per base pair of sequence alignment. The red spectrum records tract frequencies compiled from the entire alignment, while the blue spectra result from 100 repetitions of block bootstrap resampling. A slight upward concavity around
-base IBS tracts observed per base pair of sequence alignment. The red spectrum records tract frequencies compiled from the entire alignment, while the blue spectra result from 100 repetitions of block bootstrap resampling. A slight upward concavity around  base pairs is the signature of the out of Africa bottleneck in Europeans.
base pairs is the signature of the out of Africa bottleneck in Europeans.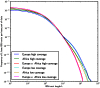
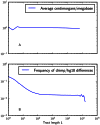
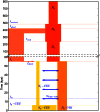
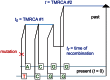
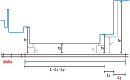
 as well as the times
as well as the times  ,
,  , and
, and  when recombinations occurred.
when recombinations occurred.References
-
- Templeton A (2002) Out of Africa again and again. Nature 416: 45–51. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

