An overview of STRUCTURE: applications, parameter settings, and supporting software
- PMID: 23755071
- PMCID: PMC3665925
- DOI: 10.3389/fgene.2013.00098
An overview of STRUCTURE: applications, parameter settings, and supporting software
Abstract
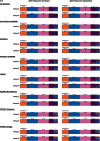
Objectives: We present an up-to-date review of STRUCTURE software: one of the most widely used population analysis tools that allows researchers to assess patterns of genetic structure in a set of samples. STRUCTURE can identify subsets of the whole sample by detecting allele frequency differences within the data and can assign individuals to those sub-populations based on analysis of likelihoods. The review covers STRUCTURE's most commonly used ancestry and frequency models, plus an overview of the main applications of the software in human genetics including case-control association studies (CCAS), population genetics, and forensic analysis. The review is accompanied by supplementary material providing a step-by-step guide to running STRUCTURE.
Methods: With reference to a worked example, we explore the effects of changing the principal analysis parameters on STRUCTURE results when analyzing a uniform set of human genetic data. Use of the supporting software: CLUMPP and distruct is detailed and we provide an overview and worked example of STRAT software, applicable to CCAS.
Conclusion: The guide offers a simplified view of how STRUCTURE, CLUMPP, distruct, and STRAT can be applied to provide researchers with an informed choice of parameter settings and supporting software when analyzing their own genetic data.
Keywords: CLUMPP; STRAT; STRUCTURE; case-control association studies; distruct; population structure; stratification.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

