Signal peptide cleavage from GP5 of PRRSV: a minor fraction of molecules retains the decoy epitope, a presumed molecular cause for viral persistence
- PMID: 23755249
- PMCID: PMC3675037
- DOI: 10.1371/journal.pone.0065548
Signal peptide cleavage from GP5 of PRRSV: a minor fraction of molecules retains the decoy epitope, a presumed molecular cause for viral persistence
Abstract
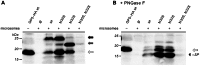
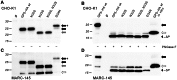
Porcine reproductive and respiratory syndrome virus (PRRSV) is the major pathogen in the pig industry. Variability of the antigens and persistence are the biggest challenges for successful control and elimination of the disease. GP5, the major glycoprotein of PRRSV, is considered an important target of neutralizing antibodies, which however appear only late in infection. This was attributed to the presence of a "decoy epitope" located near a hypervariable region of GP5. This region also harbors the predicted signal peptide cleavage sites and (dependent on the virus strain) a variable number of potential N-glycosylation sites. Molecular processing of GP5 has not been addressed experimentally so far: whether and where the signal peptide is cleaved and (as a consequence) whether the "decoy epitope" is present in virus particles. We show that the signal peptide of GP5 from the American type 2 reference strain VR-2332 is cleaved, both during in vitro translation in the presence of microsomes and in transfected cells. This was found to be independent of neighboring glycosylation sites and occurred in a variety of porcine cells for GP5 sequences derived from various type 2 strains. The exact signal peptide cleavage site was elucidated by mass spectrometry of virus-derived and recombinant GP5. The results revealed that the signal peptide of GP5 is cleaved at two sites. As a result, a mixture of GP5 proteins exists in virus particles, some of which still contain the "decoy epitope" sequence. Heterogeneity was also observed for the use of glycosylation sites in the hypervariable region. Lastly, GP5 mutants were engineered where one of the signal peptide cleavage sites was blocked. Wildtype GP5 exhibited exactly the same SDS-PAGE mobility as the mutant that is cleavable at site 2 only. This indicates that the overwhelming majority of all GP5 molecules does not contain the "decoy epitope".
Conflict of interest statement
Figures



References
-
- Snijder EJ, Meulenberg JJ (1998) The molecular biology of arteriviruses. J Gen Virol 79: 961–979. - PubMed
-
- Wensvoort G, Terpstra C, Pol JM, ter Laak EA, Bloemraad M, et al. (1991) Mystery swine disease in The Netherlands: the isolation of Lelystad virus. Vet Q 13: 121–130. - PubMed
-
- Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, et al. (1992) Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest 4: 117–126. - PubMed
-
- Zhou L, Yang H (2010) Porcine reproductive and respiratory syndrome in China. Virus Res 154: 31–37. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

