A DIStinctively novel exoribonuclease that really likes U
- PMID: 23756464
- PMCID: PMC3981174
- DOI: 10.1038/emboj.2013.136
A DIStinctively novel exoribonuclease that really likes U
Abstract
Regulated degradation plays a major role in determining the levels of both non-coding (miRNA) and coding (mRNA) transcripts. Thus, insights into the factors and pathways that influence this process have broad, interdisciplinary implications. New findings by Malecki et al (2013), Lubas et al (2013), and Chang et al (2013) identify the protein Dis3L2 as a major player in the 3′–5′ exonucleolytic decay of transcripts. Furthermore, they demonstrate a strong connection between terminal uridylation of the RNA substrate and enzymatic activity.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
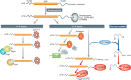
Comment on
-
The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway.EMBO J. 2013 Jul 3;32(13):1842-54. doi: 10.1038/emboj.2013.63. Epub 2013 Mar 15. EMBO J. 2013. PMID: 23503588 Free PMC article.
-
A role for the Perlman syndrome exonuclease Dis3l2 in the Lin28-let-7 pathway.Nature. 2013 May 9;497(7448):244-8. doi: 10.1038/nature12119. Epub 2013 Apr 17. Nature. 2013. PMID: 23594738 Free PMC article.
-
Exonuclease hDIS3L2 specifies an exosome-independent 3'-5' degradation pathway of human cytoplasmic mRNA.EMBO J. 2013 Jul 3;32(13):1855-68. doi: 10.1038/emboj.2013.135. Epub 2013 Jun 11. EMBO J. 2013. PMID: 23756462 Free PMC article.
References
-
- Astuti D, Morris MR, Cooper WN, Staals RH, Wake NC, Fews GA, Gill H, Gentle D, Shuib S, Ricketts CJ, Cole T, van Essen AJ, van Lingen RA, Neri G, Opitz JM, Rump P, Stolte-Dijkstra I, Müller F, Pruijn GJ, Latif F et al. (2012) Germline mutations in DIS3L2 cause the Perlman syndrome of overgrowth and Wilms tumor susceptibility. Nat Genet 44: 277–284 - PubMed
-
- Heo I, Joo C, Kim YK, Ha M, Yoon MJ, Cho J, Yeom KH, Han J, Kim VN (2009) TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre-microRNA uridylation. Nat Cell Biol 11: 1157–1163 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

