Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates
- PMID: 23758874
- PMCID: PMC3685588
- DOI: 10.1186/1471-2164-14-390
Pyrosequencing analysis of the human microbiota of healthy Chinese undergraduates
Abstract
Background: Elucidating the biogeography of bacterial communities on the human body is critical for establishing healthy baselines from which to detect differences associated with disease; however, little is known about the baseline bacterial profiles from various human habitats of healthy Chinese undergraduates.
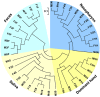
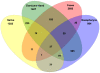
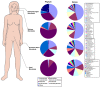
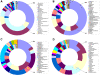
Results: Using parallel barcoded 454 pyrosequencing targeting on the 16S rRNA gene V3 region, the bacterial diversity of the nasopharynx, saliva, dominant hands, and feces were investigated from 10 healthy Chinese junior boarding undergraduates at Zhejiang University. The participants were 21-24 years of age with a body mass index (BMI) < 24 kg/m(2). A total of 156,717 high-quality pyrosequencing reads were obtained for evaluating bacterial diversity, which represented 29,887 unique phylotypes. The overall taxonomic distribution of the 16S rRNA gene-based amplicons demonstrated that these 4 habitats of the human body harbored distinct microbiota and could be divided into different clusters according to anatomic site, while the established patterns of bacterial diversity followed the human body habitat (feces, hands, saliva, and nasopharynx). Although significant inter-individual variation was observed, the healthy microbiota still shared a large number of phylotypes in each habitat, but not among the four habitats, indicating that a core microbiome existed in each healthy habitat. The vast majority of sequences from these different habitats were classified into different taxonmies that became the predominant bacteria of the healthy microbiota.
Conclusions: We first established the framework of microbial communities from four healthy human habitats of the same participants with similar living environments for the Chinese undergraduates. Our data represent an important step for determining the diversity of Chinese healthy microbiota, and can be used for more large-scale studies that focus on the interactions between healthy and diseases states for young Chinese adults in the same age range.
Figures




References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

