Utilizing protein structure to identify non-random somatic mutations
- PMID: 23758891
- PMCID: PMC3691676
- DOI: 10.1186/1471-2105-14-190
Utilizing protein structure to identify non-random somatic mutations
Abstract
Background: Human cancer is caused by the accumulation of somatic mutations in tumor suppressors and oncogenes within the genome. In the case of oncogenes, recent theory suggests that there are only a few key "driver" mutations responsible for tumorigenesis. As there have been significant pharmacological successes in developing drugs that treat cancers that carry these driver mutations, several methods that rely on mutational clustering have been developed to identify them. However, these methods consider proteins as a single strand without taking their spatial structures into account. We propose an extension to current methodology that incorporates protein tertiary structure in order to increase our power when identifying mutation clustering.
Results: We have developed iPAC (identification of Protein Amino acid Clustering), an algorithm that identifies non-random somatic mutations in proteins while taking into account the three dimensional protein structure. By using the tertiary information, we are able to detect both novel clusters in proteins that are known to exhibit mutation clustering as well as identify clusters in proteins without evidence of clustering based on existing methods. For example, by combining the data in the Protein Data Bank (PDB) and the Catalogue of Somatic Mutations in Cancer, our algorithm identifies new mutational clusters in well known cancer proteins such as KRAS and PI3KC α. Further, by utilizing the tertiary structure, our algorithm also identifies clusters in EGFR, EIF2AK2, and other proteins that are not identified by current methodology. The R package is available at: http://www.bioconductor.org/packages/2.12/bioc/html/iPAC.html.
Conclusion: Our algorithm extends the current methodology to identify oncogenic activating driver mutations by utilizing tertiary protein structure when identifying nonrandom somatic residue mutation clusters.
Figures

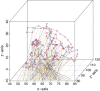


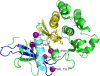


Similar articles
-
A spatial simulation approach to account for protein structure when identifying non-random somatic mutations.BMC Bioinformatics. 2014 Jul 3;15:231. doi: 10.1186/1471-2105-15-231. BMC Bioinformatics. 2014. PMID: 24990767 Free PMC article.
-
A graph theoretic approach to utilizing protein structure to identify non-random somatic mutations.BMC Bioinformatics. 2014 Mar 26;15:86. doi: 10.1186/1471-2105-15-86. BMC Bioinformatics. 2014. PMID: 24669769 Free PMC article.
-
Leveraging protein quaternary structure to identify oncogenic driver mutations.BMC Bioinformatics. 2016 Mar 22;17:137. doi: 10.1186/s12859-016-0963-3. BMC Bioinformatics. 2016. PMID: 27001666 Free PMC article.
-
[Current topics in mutations in the cancer genome].Nihon Geka Gakkai Zasshi. 2012 Mar;113(2):185-90. Nihon Geka Gakkai Zasshi. 2012. PMID: 22582578 Review. Japanese.
-
Successes and limitations of targeted cancer therapy in lung cancer.Prog Tumor Res. 2014;41:62-77. doi: 10.1159/000355902. Epub 2014 Feb 17. Prog Tumor Res. 2014. PMID: 24727987 Review.
Cited by
-
Adaptive genetics reveals constraints on protein structure/function by evolving E. coli under constant nutrient limitation.BMC Biol. 2025 Aug 20;23(1):261. doi: 10.1186/s12915-025-02331-7. BMC Biol. 2025. PMID: 40830465 Free PMC article.
-
A spatial simulation approach to account for protein structure when identifying non-random somatic mutations.BMC Bioinformatics. 2014 Jul 3;15:231. doi: 10.1186/1471-2105-15-231. BMC Bioinformatics. 2014. PMID: 24990767 Free PMC article.
-
Computational characterisation of cancer molecular profiles derived using next generation sequencing.Contemp Oncol (Pozn). 2015;19(1A):A78-91. doi: 10.5114/wo.2014.47137. Contemp Oncol (Pozn). 2015. PMID: 25691827 Free PMC article. Review.
-
Computational Methods Summarizing Mutational Patterns in Cancer: Promise and Limitations for Clinical Applications.Cancers (Basel). 2023 Mar 24;15(7):1958. doi: 10.3390/cancers15071958. Cancers (Basel). 2023. PMID: 37046619 Free PMC article. Review.
-
Systematic analysis of mutation distribution in three dimensional protein structures identifies cancer driver genes.Sci Rep. 2016 May 26;6:26483. doi: 10.1038/srep26483. Sci Rep. 2016. PMID: 27225414 Free PMC article.
References
-
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10(8):789–799. doi: 10.1038/nm1087. [ http://www.ncbi.nlm.nih.gov/pubmed/15286780] [PMID: 15286780] - DOI - PubMed
-
- Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D. et al.Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [ http://www.nature.com/doifinder/10.1038/nature05610] - DOI - DOI - PMC - PubMed
-
- Weinstein IB, Joe AK. Mechanisms of disease: Oncogene addiction–a rationale for molecular targeting in cancer therapy. Nat Clin Pract Oncol. 2006;3(8):448–457. doi: 10.1038/ncponc0558. [ http://www.ncbi.nlm.nih.gov/pubmed/16894390] [PMID: 16894390] - DOI - PubMed
-
- Torkamani A, Schork NJ. Prediction of cancer driver mutations in protein kinases. Cancer Res. 2008;68(6):1675–1682. doi: 10.1158/0008-5472.CAN-07-5283. [ http://www.ncbi.nlm.nih.gov/pubmed/18339846] [PMID: 18339846] - DOI - PubMed
-
- Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JKV, Parmigiani G, Kinzler KW, Vogelstein B, Velculescu VE. Mutational analysis of the tyrosine kinome in colorectal cancers. Sci (New York, N.Y.) 2003;300(5621):949. doi: 10.1126/science.1082596. [ http://www.ncbi.nlm.nih.gov/pubmed/12738854] [PMID: 12738854] - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

