Live imaging and analysis of postnatal mouse retinal development
- PMID: 23758927
- PMCID: PMC3698203
- DOI: 10.1186/1471-213X-13-24
Live imaging and analysis of postnatal mouse retinal development
Abstract
Background: The explanted, developing rodent retina provides an efficient and accessible preparation for use in gene transfer and pharmacological experimentation. Many of the features of normal development are retained in the explanted retina, including retinal progenitor cell proliferation, heterochronic cell production, interkinetic nuclear migration, and connectivity. To date, live imaging in the developing retina has been reported in non-mammalian and mammalian whole-mount samples. An integrated approach to rodent retinal culture/transfection, live imaging, cell tracking, and analysis in structurally intact explants greatly improves our ability to assess the kinetics of cell production.
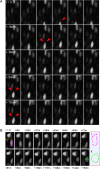
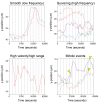
Results: In this report, we describe the assembly and maintenance of an in vitro, CO2-independent, live mouse retinal preparation that is accessible by both upright and inverted, 2-photon or confocal microscopes. The optics of this preparation permit high-quality and multi-channel imaging of retinal cells expressing fluorescent reporters for up to 48h. Tracking of interkinetic nuclear migration within individual cells, and changes in retinal progenitor cell morphology are described. Follow-up, hierarchical cluster screening revealed that several different dependent variable measures can be used to identify and group movement kinetics in experimental and control samples.
Conclusions: Collectively, these methods provide a robust approach to assay multiple features of rodent retinal development using live imaging.
Figures




Similar articles
-
Live imaging of developing mouse retinal slices.Neural Dev. 2018 Sep 15;13(1):23. doi: 10.1186/s13064-018-0120-y. Neural Dev. 2018. PMID: 30219109 Free PMC article.
-
Live Imaging of Mouse Retinal Slices.Methods Mol Biol. 2020;2092:45-53. doi: 10.1007/978-1-0716-0175-4_4. Methods Mol Biol. 2020. PMID: 31786780
-
Microglial cells in organotypic cultures of developing and adult mouse retina and their relationship with cell death.Exp Eye Res. 2014 Apr;121:42-57. doi: 10.1016/j.exer.2014.02.015. Epub 2014 Feb 27. Exp Eye Res. 2014. PMID: 24582572
-
Apoptosis in developing retinal tissue.Prog Retin Eye Res. 1999 Mar;18(2):133-65. doi: 10.1016/s1350-9462(98)00020-2. Prog Retin Eye Res. 1999. PMID: 9932281 Review.
-
Interkinetic Nuclear Migration in the Regenerating Retina.Adv Exp Med Biol. 2016;854:587-93. doi: 10.1007/978-3-319-17121-0_78. Adv Exp Med Biol. 2016. PMID: 26427463 Review.
Cited by
-
Live imaging of developing mouse retinal slices.Neural Dev. 2018 Sep 15;13(1):23. doi: 10.1186/s13064-018-0120-y. Neural Dev. 2018. PMID: 30219109 Free PMC article.
-
Culture of Adult Transgenic Zebrafish Retinal Explants for Live-cell Imaging by Multiphoton Microscopy.J Vis Exp. 2017 Feb 24;(120):55335. doi: 10.3791/55335. J Vis Exp. 2017. PMID: 28287581 Free PMC article.
-
Cephalopod retinal development shows vertebrate-like mechanisms of neurogenesis.Curr Biol. 2022 Dec 5;32(23):5045-5056.e3. doi: 10.1016/j.cub.2022.10.027. Epub 2022 Nov 9. Curr Biol. 2022. PMID: 36356573 Free PMC article.
-
Role of dynamic nuclear deformation on genomic architecture reorganization.PLoS Comput Biol. 2019 Sep 11;15(9):e1007289. doi: 10.1371/journal.pcbi.1007289. eCollection 2019 Sep. PLoS Comput Biol. 2019. PMID: 31509522 Free PMC article.
-
Nuclear envelope: positioning nuclei and organizing synapses.Curr Opin Cell Biol. 2015 Jun;34:84-93. doi: 10.1016/j.ceb.2015.06.001. Epub 2015 Jun 12. Curr Opin Cell Biol. 2015. PMID: 26079712 Free PMC article. Review.
References
-
- Donovan SL, Dyer MA. Preparation and square wave electroporation of retinal explant cultures. Nat Protoc. 2006;1(6):2710–2718. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

