Anti-microbial, anti-biofilm activities and cell selectivity of the NRC-16 peptide derived from witch flounder, Glyptocephalus cynoglossus
- PMID: 23760014
- PMCID: PMC3721208
- DOI: 10.3390/md11061836
Anti-microbial, anti-biofilm activities and cell selectivity of the NRC-16 peptide derived from witch flounder, Glyptocephalus cynoglossus
Abstract
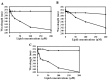
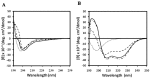
Previous studies had identified novel antimicrobial peptides derived from witch flounder. In this work, we extended the search for the activity of peptide that showed antibacterial activity on clinically isolated bacterial cells and bacterial biofilm. Pseudomonas aeruginosa was obtained from otitis media and cholelithiasis patients, while Staphylococcus aureus was isolated from otitis media patients. We found that synthetic peptide NRC-16 displays antimicrobial activity and is not sensitive to salt during its bactericidal activity. Interestingly, this peptide also led to significant inhibition of biofilm formation at a concentration of 4-16 μM. NRC-16 peptide is able to block biofilm formation at concentrations just above its minimum inhibitory concentration while conventional antibiotics did not inhibit the biofilm formation except ciprofloxacin and piperacillin. It did not cause significant lysis of human RBC, and is not cytotoxic to HaCaT cells and RAW264.7 cells, thereby indicating its selective antimicrobial activity. In addition, the peptide's binding and permeation activities were assessed by tryptophan fluorescence, calcein leakage and circular dichroism using model mammalian membranes composed of phosphatidylcholine (PC), PC/cholesterol (CH) and PC/sphingomyelin (SM). These experiments confirmed that NRC-16 does not interact with any of the liposomes but the control peptide melittin did. Taken together, we found that NRC-16 has potent antimicrobial and antibiofilm activities with less cytotoxicity, and thus can be considered for treatment of microbial infection in the future.
Figures


Similar articles
-
Design and membrane-disruption mechanism of charge-enriched AMPs exhibiting cell selectivity, high-salt resistance, and anti-biofilm properties.Amino Acids. 2016 Feb;48(2):505-22. doi: 10.1007/s00726-015-2104-0. Epub 2015 Oct 8. Amino Acids. 2016. PMID: 26450121
-
Intragenic Antimicrobial Peptide Hs02 Hampers the Proliferation of Single- and Dual-Species Biofilms of P. aeruginosa and S. aureus: A Promising Agent for Mitigation of Biofilm-Associated Infections.Int J Mol Sci. 2019 Jul 23;20(14):3604. doi: 10.3390/ijms20143604. Int J Mol Sci. 2019. PMID: 31340580 Free PMC article.
-
Melittin and its potential in the destruction and inhibition of the biofilm formation by Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa isolated from bovine milk.Microb Pathog. 2017 Nov;112:57-62. doi: 10.1016/j.micpath.2017.09.046. Epub 2017 Sep 21. Microb Pathog. 2017. PMID: 28943153
-
Antibacterial, anti-biofilm and in vivo activities of the antimicrobial peptides P5 and P6.2.Microb Pathog. 2020 Feb;139:103886. doi: 10.1016/j.micpath.2019.103886. Epub 2019 Nov 25. Microb Pathog. 2020. PMID: 31778756
-
Pse-T2, an Antimicrobial Peptide with High-Level, Broad-Spectrum Antimicrobial Potency and Skin Biocompatibility against Multidrug-Resistant Pseudomonas aeruginosa Infection.Antimicrob Agents Chemother. 2018 Nov 26;62(12):e01493-18. doi: 10.1128/AAC.01493-18. Print 2018 Dec. Antimicrob Agents Chemother. 2018. PMID: 30323036 Free PMC article.
Cited by
-
Calculation of the Global and Local Conceptual DFT Indices for the Prediction of the Chemical Reactivity Properties of Papuamides A-F Marine Drugs.Molecules. 2019 Sep 11;24(18):3312. doi: 10.3390/molecules24183312. Molecules. 2019. PMID: 31514433 Free PMC article.
-
Inhibitory Effects of Antimicrobial Peptides on Lipopolysaccharide-Induced Inflammation.Mediators Inflamm. 2015;2015:167572. doi: 10.1155/2015/167572. Epub 2015 Nov 3. Mediators Inflamm. 2015. PMID: 26612970 Free PMC article. Review.
-
How Melittin Inserts into Cell Membrane: Conformational Changes, Inter-Peptide Cooperation, and Disturbance on the Membrane.Molecules. 2019 May 7;24(9):1775. doi: 10.3390/molecules24091775. Molecules. 2019. PMID: 31067828 Free PMC article. Review.
-
Antimicrobial Peptides Design Using Deep Learning and Rational Modifications: Activity in Bacteria, Candida albicans, and Cancer Cells.Curr Microbiol. 2025 Jul 11;82(9):379. doi: 10.1007/s00284-025-04346-3. Curr Microbiol. 2025. PMID: 40643674 Free PMC article.
-
Short antimicrobial peptides as cosmetic ingredients to deter dermatological pathogens.Appl Microbiol Biotechnol. 2015 Nov;99(21):8847-55. doi: 10.1007/s00253-015-6926-1. Epub 2015 Aug 26. Appl Microbiol Biotechnol. 2015. PMID: 26307444 Free PMC article. Review.
References
-
- Dzidic S., Suskovic J., Kos B. Antibiotic resistance mechanisms in bacteria: Biochemical and genetic aspects. Food Technol. Biotechnol. 2008;46:11–21.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

