Missense mutations of MLH1 and MSH2 genes detected in patients with gastrointestinal cancer are associated with exonic splicing enhancers and silencers
- PMID: 23760103
- PMCID: PMC3678577
- DOI: 10.3892/ol.2013.1243
Missense mutations of MLH1 and MSH2 genes detected in patients with gastrointestinal cancer are associated with exonic splicing enhancers and silencers
Abstract
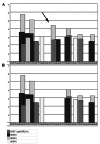
The MLH1 and MSH2 genes in DNA mismatch repair are important in the pathogenesis of gastrointestinal cancer. Recent studies of normal and alternative splicing suggest that the deleterious effects of missense mutations may in fact be splicing-related when they are located in exonic splicing enhancers (ESEs) or exonic splicing silencers (ESSs). In this study, we used ESE-finder and FAS-ESS software to analyze the potential ESE/ESS motifs of the 114 missense mutations detected in the two genes in East Asian gastrointestinal cancer patients. In addition, we used the SIFT tool to functionally analyze these mutations. The amount of the ESE losses (68) was 51.1% higher than the ESE gains (45) of all the mutations. However, the amount of the ESS gains (27) was 107.7% higher than the ESS losses (13). In total, 56 (49.1%) mutations possessed a potential exonic splicing regulator (ESR) error. Eighty-one mutations (71.1%) were predicted to be deleterious with a lower tolerance index as detected by the Sorting Intolerant from Tolerant (SIFT) tool. Among these, 38 (33.3%) mutations were predicted to be functionally deleterious and possess one potential ESR error, while 18 (15.8%) mutations were predicted to be functionally deleterious and exhibit two potential ESR errors. These may be more likely to affect exon splicing. Our results indicated that there is a strong correlation between missense mutations in MLH1 and MSH2 genes detected in East Asian gastrointestinal cancer patients and ESR motifs. In order to correctly understand the molecular nature of mutations, splicing patterns should be compared between wild-type and mutant samples.
Keywords: MLH1; MSH2; exonic splicing enhancer; exonic splicing silencer; gastrointestinal cancer; missense mutation.
Figures
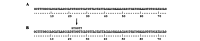
Similar articles
-
Systematic mRNA analysis for the effect of MLH1 and MSH2 missense and silent mutations on aberrant splicing.Hum Mutat. 2006 Feb;27(2):145-54. doi: 10.1002/humu.20280. Hum Mutat. 2006. PMID: 16395668
-
Missense mutations in cancer suppressor gene TP53 are colocalized with exonic splicing enhancers (ESEs).Mutat Res. 2004 Oct 4;554(1-2):175-83. doi: 10.1016/j.mrfmmm.2004.04.014. Mutat Res. 2004. PMID: 15450416
-
A large fraction of unclassified variants of the mismatch repair genes MLH1 and MSH2 is associated with splicing defects.Hum Mutat. 2008 Dec;29(12):1412-24. doi: 10.1002/humu.20796. Hum Mutat. 2008. PMID: 18561205
-
Aberrant splicing in MLH1 and MSH2 due to exonic and intronic variants.Hum Genet. 2006 Mar;119(1-2):9-22. doi: 10.1007/s00439-005-0107-8. Epub 2005 Dec 8. Hum Genet. 2006. PMID: 16341550
-
Role of viral splicing elements and cellular RNA binding proteins in regulation of HIV-1 alternative RNA splicing.Curr HIV Res. 2006 Jan;4(1):43-55. doi: 10.2174/157016206775197655. Curr HIV Res. 2006. PMID: 16454710 Review.
Cited by
-
Novel Genetic Markers for Early Detection of Elevated Breast Cancer Risk in Women.Int J Mol Sci. 2019 Sep 28;20(19):4828. doi: 10.3390/ijms20194828. Int J Mol Sci. 2019. PMID: 31569399 Free PMC article.
-
Polyethylene Glycol Electrolyte Lavage Solution versus Colonic Hydrotherapy for Bowel Preparation before Colonoscopy: A Single Center, Randomized, and Controlled Study.Gastroenterol Res Pract. 2014;2014:541586. doi: 10.1155/2014/541586. Epub 2014 Jun 5. Gastroenterol Res Pract. 2014. PMID: 24995014 Free PMC article.
-
A single-base change at a splice site in Wx-A1 caused incorrect RNA splicing and gene inactivation in a wheat EMS mutant line.Theor Appl Genet. 2019 Jul;132(7):2097-2109. doi: 10.1007/s00122-019-03340-1. Epub 2019 Apr 16. Theor Appl Genet. 2019. PMID: 30993362
-
Alternative splicing of DNA damage response genes and gastrointestinal cancers.World J Gastroenterol. 2014 Dec 14;20(46):17305-13. doi: 10.3748/wjg.v20.i46.17305. World J Gastroenterol. 2014. PMID: 25516641 Free PMC article. Review.
References
-
- Peltomäki P, Vasen HF. Mutations predisposing to hereditary nonpolyposis colorectal cancer: database and results of a collaborative study. The International Collaborative Group on hereditary nonpolyposis colorectal cancer. Gastroenterology. 1997;113:1146–1158. - PubMed
-
- Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet. 2002;3:285–298. - PubMed
-
- Pagani F, Baralle FE. Genomic variants in exons and introns: identifying the splicing spoilers. Nat Rev Genet. 2004;5:389–396. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
