A simple three-dimensional-focusing, continuous-flow mixer for the study of fast protein dynamics
- PMID: 23760106
- PMCID: PMC3733270
- DOI: 10.1039/c3lc50497b
A simple three-dimensional-focusing, continuous-flow mixer for the study of fast protein dynamics
Abstract
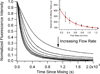
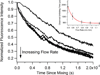
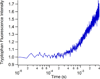
We present a simple, yet flexible microfluidic mixer with a demonstrated mixing time as short as 80 μs that is widely accessible because it is made of commercially available parts. To simplify the study of fast protein dynamics, we have developed an inexpensive continuous-flow microfluidic mixer, requiring no specialized equipment or techniques. The mixer uses three-dimensional, hydrodynamic focusing of a protein sample stream by a surrounding sheath solution to achieve rapid diffusional mixing between the sample and sheath. Mixing initiates the reaction of interest. Reactions can be spatially observed by fluorescence or absorbance spectroscopy. We characterized the pixel-to-time calibration and diffusional mixing experimentally. We achieved a mixing time as short as 80 μs. We studied the kinetics of horse apomyoglobin (apoMb) unfolding from the intermediate (I) state to its completely unfolded (U) state, induced by a pH jump from the initial pH of 4.5 in the sample stream to a final pH of 2.0 in the sheath solution. The reaction time was probed using the fluorescence of 1-anilinonaphthalene-8-sulfonate (1,8-ANS) bound to the folded protein. We observed unfolding of apoMb within 760 μs, without populating additional intermediate states under these conditions. We also studied the reaction kinetics of the conversion of pyruvate to lactate catalyzed by lactate dehydrogenase using the intrinsic tryptophan emission of the enzyme. We observe sub-millisecond kinetics that we attribute to Michaelis complex formation and loop domain closure. These results demonstrate the utility of the three-dimensional focusing mixer for biophysical studies of protein dynamics.
Figures


Similar articles
-
Dynamics of ANS binding to tuna apomyoglobin measured with fluorescence correlation spectroscopy.Biophys J. 2001 Dec;81(6):3510-21. doi: 10.1016/S0006-3495(01)75982-6. Biophys J. 2001. PMID: 11721012 Free PMC article.
-
Analysis of heterogeneous fluorescence decays in proteins. Using fluorescence lifetime of 8-anilino-1-naphthalenesulfonate to probe apomyoglobin unfolding at equilibrium.Biochim Biophys Acta. 2006 Jul;1760(7):1125-37. doi: 10.1016/j.bbagen.2006.02.019. Epub 2006 Mar 31. Biochim Biophys Acta. 2006. PMID: 16730413
-
Microsecond folding dynamics of apomyoglobin at acidic pH.J Phys Chem B. 2012 Jun 14;116(23):7014-25. doi: 10.1021/jp3012365. Epub 2012 Apr 17. J Phys Chem B. 2012. PMID: 22475221 Free PMC article.
-
Folding of apomyoglobin: Analysis of transient intermediate structure during refolding using quick hydrogen deuterium exchange and NMR.Proc Jpn Acad Ser B Phys Biol Sci. 2017;93(1):10-27. doi: 10.2183/pjab.93.002. Proc Jpn Acad Ser B Phys Biol Sci. 2017. PMID: 28077807 Free PMC article. Review.
-
The folding process of apomyoglobin.Protein Pept Lett. 2005 Apr;12(3):229-34. doi: 10.2174/0929866053587174. Protein Pept Lett. 2005. PMID: 15777270 Review.
Cited by
-
Submillisecond mixing in a continuous-flow, microfluidic mixer utilizing mid-infrared hyperspectral imaging detection.Lab Chip. 2014 Feb 7;14(3):584-91. doi: 10.1039/c3lc51171e. Lab Chip. 2014. PMID: 24302515 Free PMC article.
-
An Efficient 3D-Printed Gravity Mixer for Lab-on-a-CD Applications.Micromachines (Basel). 2024 Feb 20;15(3):291. doi: 10.3390/mi15030291. Micromachines (Basel). 2024. PMID: 38542538 Free PMC article.
-
Micro total analysis systems: fundamental advances and biological applications.Anal Chem. 2014 Jan 7;86(1):95-118. doi: 10.1021/ac403688g. Epub 2013 Dec 13. Anal Chem. 2014. PMID: 24274655 Free PMC article. Review. No abstract available.
-
Sandwich-format 3D printed microfluidic mixers: a flexible platform for multi-probe analysis.J Micromech Microeng. 2015 Dec;25(12):124002. doi: 10.1088/0960-1317/25/12/124002. Epub 2015 Oct 27. J Micromech Microeng. 2015. PMID: 26855478 Free PMC article.
-
An "off-the-shelf" capillary microfluidic device that enables tuning of the droplet breakup regime at constant flow rates.Lab Chip. 2013 Dec 7;13(23):4507-11. doi: 10.1039/c3lc50804h. Lab Chip. 2013. PMID: 24122050 Free PMC article.
References
-
- Callender RH, Dyer RB. R. Gilmanshin and W. H. Woodruff. Annu Rev Phys Chem. 1998;49:173–202. - PubMed
-
- Eaton WA, Thompson PA, Chan CK. S. J. Hagen and J. Hofrichter. Structure. 1996;4:1133–1139. - PubMed
-
- Dyer RB. Curr Opin Struct Biol. 2007;17:38–47. - PubMed
-
- Wang C, Ye D-K, Wang Y-Y. T. Lu and X.-H. Xia. Lab Chip. 2013;13:1546–1553. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

