Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation
- PMID: 23760247
- PMCID: PMC3754048
- DOI: 10.1128/JVI.01216-13
Merkel cell polyomavirus large T antigen disrupts host genomic integrity and inhibits cellular proliferation
Abstract
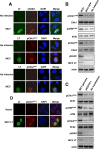
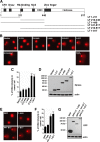
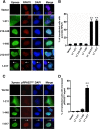
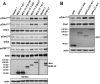
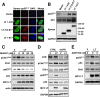
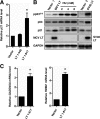
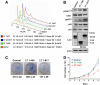
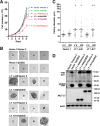
Clonal integration of Merkel cell polyomavirus (MCV) DNA into the host genome has been observed in at least 80% of Merkel cell carcinoma (MCC). The integrated viral genome typically carries mutations that truncate the C-terminal DNA binding and helicase domains of the MCV large T antigen (LT), suggesting a selective pressure to remove this MCV LT region during tumor development. In this study, we show that MCV infection leads to the activation of host DNA damage responses (DDR). This activity was mapped to the C-terminal helicase-containing region of the MCV LT. The MCV LT-activated DNA damage kinases, in turn, led to enhanced p53 phosphorylation, upregulation of p53 downstream target genes, and cell cycle arrest. Compared to the N-terminal MCV LT fragment that is usually preserved in mutants isolated from MCC tumors, full-length MCV LT shows a decreased potential to support cellular proliferation, focus formation, and anchorage-independent cell growth. These apparently antitumorigenic effects can be reversed by a dominant-negative p53 inhibitor. Our results demonstrate that MCV LT-induced DDR activates p53 pathway, leading to the inhibition of cellular proliferation. This study reveals a key difference between MCV LT and simian vacuolating virus 40 LT, which activates a DDR but inhibits p53 function. This study also explains, in part, why truncation mutations that remove the MCV LT C-terminal region are necessary for the oncogenic progression of MCV-associated cancers.
Figures

References
-
- Gjoerup O, Chang Y. 2010. Update on human polyomaviruses and cancer. Adv. Cancer Res. 106:1–51 - PubMed
-
- Kassem A, Schöpflin A, Diaz C, Weyers W, Stickeler E, Werner M, Zur Hausen A. 2008. Frequent detection of Merkel cell polyomavirus in human Merkel cell carcinomas and identification of a unique deletion in the VP1 gene. Cancer Res. 68:5009–5013 - PubMed
-
- Rodig SJ, Cheng J, Wardzala J, DoRosario A, Scanlon JJ, Laga AC, Martinez-Fernandez A, Barletta JA, Bellizzi AM, Sadasivam S, Holloway DT, Cooper DJ, Kupper TS, Wang LC, DeCaprio JA. 2012. Improved detection suggests all Merkel cell carcinomas harbor Merkel polyomavirus. J. Clin. Invest. 122:4645–4653 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

