Scaling down DNA circuits with competitive neural networks
- PMID: 23760296
- PMCID: PMC4043154
- DOI: 10.1098/rsif.2013.0212
Scaling down DNA circuits with competitive neural networks
Abstract
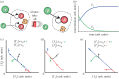
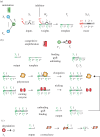
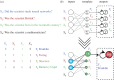
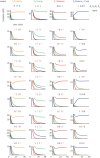
DNA has proved to be an exquisite substrate to compute at the molecular scale. However, nonlinear computations (such as amplification, comparison or restoration of signals) remain costly in term of strands and are prone to leak. Kim et al. showed how competition for an enzymatic resource could be exploited in hybrid DNA/enzyme circuits to compute a powerful nonlinear primitive: the winner-take-all (WTA) effect. Here, we first show theoretically how the nonlinearity of the WTA effect allows the robust and compact classification of four patterns with only 16 strands and three enzymes. We then generalize this WTA effect to DNA-only circuits and demonstrate similar classification capabilities with only 23 strands.
Keywords: molecular programming; pattern recognition; strand displacement circuits; winner-take-all.
Figures



References
-
- Benenson Y. 2009. Biocomputers: from test tubes to live cells. Mol. Biosyst. 5, 675–68510.1039/b902484k (doi:10.1039/b902484k) - DOI - DOI - PMC - PubMed
-
- Nandagopal N, Elowitz MB. 2011. Synthetic biology: integrated gene circuits. Science 333, 1244–124810.1126/science.1207084 (doi:10.1126/science.1207084) - DOI - DOI - PMC - PubMed
-
- Hockenberry AJ, Jewett MC. 2012. Synthetic in vitro circuits. Curr. Opin. Chem. Biol. 16, 253–25910.1016/j.cbpa.2012.05.179 (doi:10.1016/j.cbpa.2012.05.179) - DOI - DOI - PMC - PubMed
-
- Rothemund PWK. 2006. Folding DNA to create nanoscale shapes and patterns. Nature 440, 297–30210.1038/nature04586 (doi:10.1038/nature04586) - DOI - DOI - PubMed
-
- Douglas SM, Dietz H, Liedl T, Hogberg B, Graf F, Shih WM. 2009. Self-assembly of DNA into nanoscale three-dimensional shapes. Nature 459, 414–41810.1038/nature08016 (doi:10.1038/nature08016) - DOI - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
