Age-dependent stochastic models for understanding population fluctuations in continuously cultured cells
- PMID: 23760298
- PMCID: PMC3971721
- DOI: 10.1098/rsif.2013.0325
Age-dependent stochastic models for understanding population fluctuations in continuously cultured cells
Abstract
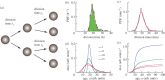
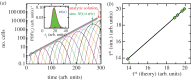
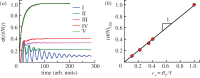
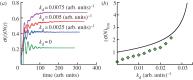
For symmetrically dividing cells, large variations in the cell cycle time are typical, even among clonal cells. The consequence of this variation is important in stem cell differentiation, tissue and organ size control, and cancer development, where cell division rates ultimately determine the cell population. We explore the connection between cell cycle time variation and population-level fluctuations using simple stochastic models. We find that standard population models with constant division and death rates fail to predict the level of population fluctuation. Instead, variations in the cell division time contribute to population fluctuations. An age-dependent birth and death model allows us to compute the mean squared fluctuation or the population dispersion as a function of time. This dispersion grows exponentially with time, but scales with the population. We also find a relationship between the dispersion and the cell cycle time distribution for synchronized cell populations. The model can easily be generalized to study populations involving cell differentiation and competitive growth situations.
Keywords: cell cycle variation; evolutionary models; stochastic population dynamics.
Figures

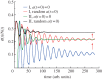
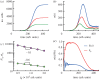
References
-
- Wang P, Robert L, Pelletier J, Dang WL, Taddei F, Wright A, Jun S. 2010. Robust growth of Escherichia coli. Curr. Biol. 20, 1099–110310.1016/j.cub.2010.04.045 (doi:10.1016/j.cub.2010.04.045) - DOI - DOI - PMC - PubMed
-
- Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. 2009. Cell growth and size homeostasis in proliferating animal cells. Science 325, 167–17110.1126/science.1174294 (doi:10.1126/science.1174294) - DOI - DOI - PMC - PubMed
-
- Halter M, Elliott JT, Hubbard JB, Tona A, Plant AL. 2009. Cell colume distributions reveal cell growth and division times. J. Theor. Biol. 257, 124–13010.1016/j.jtbi.2008.10.031 (doi:10.1016/j.jtbi.2008.10.031) - DOI - DOI - PubMed
-
- Scott M, Gunderson CW, Mateescu EM, Zhang Z, Hwa T. 2010. Interdependence of cell growth and gene expression: origins and consequences. Science 330, 1099–110210.1126/science.1192588 (doi:10.1126/science.1192588) - DOI - DOI - PubMed
-
- Sun Z, Komarova NL. 2012. Stochastic modeling of stem-cell dynamics with control. Math. Biosci. 240, 231–24010.1016/j.mbs.2012.08.004 (doi:10.1016/j.mbs.2012.08.004) - DOI - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

