An out-of-frame overlapping reading frame in the ataxin-1 coding sequence encodes a novel ataxin-1 interacting protein
- PMID: 23760502
- PMCID: PMC3724639
- DOI: 10.1074/jbc.M113.472654
An out-of-frame overlapping reading frame in the ataxin-1 coding sequence encodes a novel ataxin-1 interacting protein
Abstract
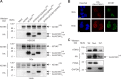
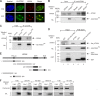
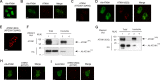
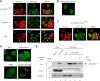
Spinocerebellar ataxia type 1 is an autosomal dominant cerebellar ataxia associated with the expansion of a polyglutamine tract within the ataxin-1 (ATXN1) protein. Recent studies suggest that understanding the normal function of ATXN1 in cellular processes is essential to decipher the pathogenesis mechanisms in spinocerebellar ataxia type 1. We found an alternative translation initiation ATG codon in the +3 reading frame of human ATXN1 starting 30 nucleotides downstream of the initiation codon for ATXN1 and ending at nucleotide 587. This novel overlapping open reading frame (ORF) encodes a 21-kDa polypeptide termed Alt-ATXN1 (Alternative ATXN1) with a completely different amino acid sequence from ATXN1. We introduced a hemagglutinin tag in-frame with Alt-ATXN1 in ATXN1 cDNA and showed in cell culture the co-expression of both ATXN1 and Alt-ATXN1. Remarkably, Alt-ATXN1 colocalized and interacted with ATXN1 in nuclear inclusions. In contrast, in the absence of ATXN1 expression, Alt-ATXN1 displays a homogenous nucleoplasmic distribution. Alt-ATXN1 interacts with poly(A)(+) RNA, and its nuclear localization is dependent on RNA transcription. Polyclonal antibodies raised against Alt-ATXN1 confirmed the expression of Alt-ATXN1 in human cerebellum expressing ATXN1. These results demonstrate that human ATXN1 gene is a dual coding sequence and that ATXN1 interacts with and controls the subcellular distribution of Alt-ATXN1.
Keywords: Alternative Translation Initiation; Ataxia; Dual Coding Gene; Gene Expression; Nucleus; Overlapping Reading Frame; RNA-Binding Protein; Spinocerebellar Ataxia Type 1; Translation.
Figures



References
-
- Orr H. T., Chung M. Y., Banfi S., Kwiatkowski T. J., Jr, Servadio A., Beaudet A. L., McCall A. E., Duvick L. A., Ranum L. P., Zoghbi H. Y. (1993) Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat. Genet. 4, 221–226 - PubMed
-
- Chung M. Y., Ranum L. P., Duvick L. A., Servadio A., Zoghbi H. Y., Orr H. T. (1993) Evidence for a mechanism predisposing to intergenerational CAG repeat instability in spinocerebellar ataxia type I. Nat. Genet. 5, 254–258 - PubMed
-
- Cummings C. J., Mancini M. A., Antalffy B., DeFranco D. B., Orr H. T., Zoghbi H. Y. (1998) Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat. Genet. 19, 148–154 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

