Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p
- PMID: 23760605
- PMCID: PMC3672780
- DOI: 10.3389/fcimb.2013.00019
Actin-binding protein regulation by microRNAs as a novel microbial strategy to modulate phagocytosis by host cells: the case of N-Wasp and miR-142-3p
Abstract
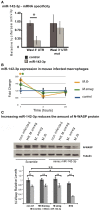
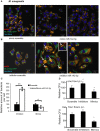
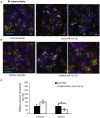
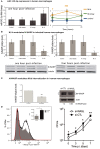
Mycobacterium tuberculosis (Mtb) is a successful intracellular pathogen that thrives in macrophages (Mφs). There is a need to better understand how Mtb alters cellular processes like phagolysosome biogenesis, a classical determinant of its pathogenesis. A central feature of this bacteria's strategy is the manipulation of Mφ actin. Here, we examined the role of microRNAs (miRNAs) as a potential mechanism in the regulation of actin-mediated events leading to phagocytosis in the context of mycobacteria infection. Given that non-virulent Mycobacterium smegmatis also controls actin filament assembly to prolong its intracellular survival inside host cells, we performed a global transcriptomic analysis to assess the modulation of miRNAs upon M. smegmatis infection of the murine Mφ cell line, J774A.1. This approach identified miR-142-3p as a key candidate to be involved in the regulation of actin dynamics required in phagocytosis. We unequivocally demonstrate that miR-142-3p targets N-Wasp, an actin-binding protein required during microbial challenge. A gain-of-function approach for miR-142-3p revealed a down-regulation of N-Wasp expression accompanied by a decrease of mycobacteria intake, while a loss-of-function approach yielded the reciprocal increase of the phagocytosis process. Equally important, we show Mtb induces the early expression of miR-142-3p and partially down-regulates N-Wasp protein levels in both the murine J774A.1 cell line and primary human Mφs. As proof of principle, the partial siRNA-mediated knock down of N-Wasp resulted in a decrease of Mtb intake by human Mφs, reflected in lower levels of colony-forming units (CFU) counts over time. We therefore propose the modulation of miRNAs as a novel strategy in mycobacterial infection to control factors involved in actin filament assembly and other early events of phagolysosome biogenesis.
Keywords: M. smegmatis; M. tuberculosis; N-Wasp; macrophage; miR-142-3p; miRNA; phagocytosis; tuberculosis.
Figures

References
-
- Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008). The microRNA.org resource: targets and expression. Nucleic Acids Res. 36, D149–D153 10.1093/nar/gkm995 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

