Analysis of stress-responsive transcriptome in the intestine of Asian seabass (Lates calcarifer) using RNA-seq
- PMID: 23761194
- PMCID: PMC3789556
- DOI: 10.1093/dnares/dst022
Analysis of stress-responsive transcriptome in the intestine of Asian seabass (Lates calcarifer) using RNA-seq
Abstract
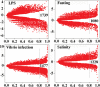
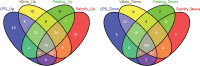
Identification of differentially expressed genes (DEGs) and regulated pathways in response to stressors using a whole-genome approach is critical to understanding the mechanisms underlying stress responses. We challenged Asian seabass with lipopolysaccharide (LPS), Vibrio harveyi, high salinity and fasting, and sequenced six cDNA libraries of intestine samples using Roche 454 RNA-seq. Over 1 million reads (average size: 516 bp) were obtained. The de novo assembly obtained 83 911 unisequences with an average length of 747 bp. In total, 62.3% of the unisequences were annotated. We observed overall similar expression profiles among different challenges, while a number of DEGs and regulated pathways were identified under specific challenges. More than 1000 DEGs and over 200 regulated pathways for each stressor were identified. Thirty-seven genes were differentially expressed in response to all challenges. Our data suggest that there is a global coordination and fine-tuning of gene regulation during different challenges. In addition, we detected dramatic immune responses in intestines under different stressors. This study is the first step towards the comprehensive understanding of the mechanisms underlying stress responses and supplies significant transcriptome resources for studying biological questions in non-model fish species.
Keywords: RNA-seq; disease; intestine; nutrition; stress.
Figures


Similar articles
-
RNA-Seq analysis of salinity stress-responsive transcriptome in the liver of spotted sea bass (Lateolabrax maculatus).PLoS One. 2017 Mar 2;12(3):e0173238. doi: 10.1371/journal.pone.0173238. eCollection 2017. PLoS One. 2017. PMID: 28253338 Free PMC article.
-
Transcriptome analysis of genes responding to NNV infection in Asian seabass epithelial cells.Fish Shellfish Immunol. 2016 Jul;54:342-52. doi: 10.1016/j.fsi.2016.04.029. Epub 2016 Apr 21. Fish Shellfish Immunol. 2016. PMID: 27109582
-
Analysis of the Asian seabass transcriptome based on expressed sequence tags.DNA Res. 2011 Dec;18(6):513-22. doi: 10.1093/dnares/dsr036. Epub 2011 Nov 15. DNA Res. 2011. PMID: 22086997 Free PMC article.
-
Identification and characterization of 63 MicroRNAs in the Asian seabass Lates calcarifer.PLoS One. 2011 Mar 11;6(3):e17537. doi: 10.1371/journal.pone.0017537. PLoS One. 2011. PMID: 21412421 Free PMC article.
-
Comparative Study of Immune Reaction Against Bacterial Infection From Transcriptome Analysis.Front Immunol. 2019 Feb 5;10:153. doi: 10.3389/fimmu.2019.00153. eCollection 2019. Front Immunol. 2019. PMID: 30804945 Free PMC article. Review.
Cited by
-
Small-scale transcriptomics reveals differences among gonadal stages in Asian seabass (Lates calcarifer).Reprod Biol Endocrinol. 2014 Jan 9;12:5. doi: 10.1186/1477-7827-12-5. Reprod Biol Endocrinol. 2014. PMID: 24405829 Free PMC article.
-
Freshwater transfer affected intestinal microbiota with correlation to cytokine gene expression in Asian sea bass.Front Microbiol. 2023 Apr 6;14:1097954. doi: 10.3389/fmicb.2023.1097954. eCollection 2023. Front Microbiol. 2023. PMID: 37089546 Free PMC article.
-
Scale Drop Disease Virus (SDDV) and Lates calcarifer Herpes Virus (LCHV) Coinfection Downregulate Immune-Relevant Pathways and Cause Splenic and Kidney Necrosis in Barramundi Under Commercial Farming Conditions.Front Genet. 2021 Jun 18;12:666897. doi: 10.3389/fgene.2021.666897. eCollection 2021. Front Genet. 2021. PMID: 34220943 Free PMC article.
-
Biochemical indices, gene expression, and SNPs associated with salinity adaptation in juvenile chum salmon (Oncorhynchus keta) as determined by comparative transcriptome analysis.PeerJ. 2022 Sep 12;10:e13585. doi: 10.7717/peerj.13585. eCollection 2022. PeerJ. 2022. PMID: 36117540 Free PMC article.
-
The Change of Teleost Skin Commensal Microbiota Is Associated With Skin Mucosal Transcriptomic Responses During Parasitic Infection by Ichthyophthirius multifillis.Front Immunol. 2018 Dec 18;9:2972. doi: 10.3389/fimmu.2018.02972. eCollection 2018. Front Immunol. 2018. PMID: 30619329 Free PMC article.
References
-
- Roelofs D., Aarts M.G.M., Schat H., van Straalen N.M. Functional ecological genomics to demonstrate general and specific responses to abiotic stress. Funct. Ecol. 2008;22:8–18.
-
- Harper C., Wolf J.C. Morphologic effects of the stress response in fish. ILAR J. 2009;50:387–96. - PubMed
-
- Barton B.A. Stress in fishes: a diversity of responses with particular reference to changes in circulating corticosteroids. Integ. Comp. Biol. 2002;42:517–25. - PubMed
-
- von Borell H. The biology of stress and its application to livestock housing and transportation assessment. J. Anim. Sci. 2001;79(E. Suppl.):E260–7.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

