Genomic analysis of coxsackieviruses A1, A19, A22, enteroviruses 113 and 104: viruses representing two clades with distinct tropism within enterovirus C
- PMID: 23761409
- PMCID: PMC3749054
- DOI: 10.1099/vir.0.053462-0
Genomic analysis of coxsackieviruses A1, A19, A22, enteroviruses 113 and 104: viruses representing two clades with distinct tropism within enterovirus C
Abstract
Coxsackieviruses (CV) A1, CV-A19 and CV-A22 have historically comprised a distinct phylogenetic clade within Enterovirus (EV) C. Several novel serotypes that are genetically similar to these three viruses have been recently discovered and characterized. Here, we report the coding sequence analysis of two genotypes of a previously uncharacterized serotype EV-C113 from Bangladesh and demonstrate that it is most similar to CV-A22 and EV-C116 within the capsid region. We sequenced novel genotypes of CV-A1, CV-A19 and CV-A22 from Bangladesh and observed a high rate of recombination within this group. We also report genomic analysis of the rarely reported EV-C104 circulating in the Gambia in 2009. All available EV-C104 sequences displayed a high degree of similarity within the structural genes but formed two clusters within the non-structural genes. One cluster included the recently reported EV-C117, suggesting an ancestral recombination between these two serotypes. Phylogenetic analysis of all available complete genome sequences indicated the existence of two subgroups within this distinct Enterovirus C clade: one has been exclusively recovered from gastrointestinal samples, while the other cluster has been implicated in respiratory disease.
Figures
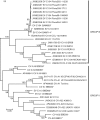


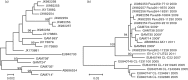
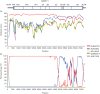
References
-
- Begier E. M., Oberste M. S., Landry M. L., Brennan T., Mlynarski D., Mshar P. A., Frenette K., Rabatsky-Ehr T., Purviance K. & other authors (2008). An outbreak of concurrent echovirus 30 and coxsackievirus A1 infections associated with sea swimming among a group of travelers to Mexico. Clin Infect Dis 47, 616–623 10.1086/590562 - DOI - PubMed
-
- Benschop K., Minnaar R., Koen G., van Eijk H., Dijkman K., Westerhuis B., Molenkamp R., Wolthers K. (2010). Detection of human enterovirus and human parechovirus (HPeV) genotypes from clinical stool samples: polymerase chain reaction and direct molecular typing, culture characteristics, and serotyping. Diagn Microbiol Infect Dis 68, 166–173 10.1016/j.diagmicrobio.2010.05.016 - DOI - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

