Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells
- PMID: 23761442
- PMCID: PMC3753646
- DOI: 10.1093/nar/gkt528
Visualization of multivalent histone modification in a single cell reveals highly concerted epigenetic changes on differentiation of embryonic stem cells
Abstract
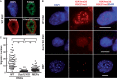
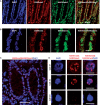
Combinations of histone modifications have significant biological roles, such as maintenance of pluripotency and cancer development, but cannot be analyzed at the single cell level. Here, we visualized a combination of histone modifications by applying the in situ proximity ligation assay, which detects two proteins in close vicinity (∼30 nm). The specificity of the method [designated as imaging of a combination of histone modifications (iChmo)] was confirmed by positive signals from H3K4me3/acetylated H3K9, H3K4me3/RNA polymerase II and H3K9me3/H4K20me3, and negative signals from H3K4me3/H3K9me3. Bivalent modification was clearly visualized by iChmo in wild-type embryonic stem cells (ESCs) known to have it, whereas rarely in Suz12 knockout ESCs and mouse embryonic fibroblasts known to have little of it. iChmo was applied to analysis of epigenetic and phenotypic changes of heterogeneous cell population, namely, ESCs at an early stage of differentiation, and this revealed that the bivalent modification disappeared in a highly concerted manner, whereas phenotypic differentiation proceeded with large variations among cells. Also, using this method, we were able to visualize a combination of repressive histone marks in tissue samples. The application of iChmo to samples with heterogeneous cell population and tissue samples is expected to clarify unknown biological and pathological significance of various combinations of epigenetic modifications.
Figures


References
-
- Meissner A. Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 2010;28:1079–1088. - PubMed
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

