Mechanisms of muscle gene regulation in the electric organ of Sternopygus macrurus
- PMID: 23761472
- PMCID: PMC3680507
- DOI: 10.1242/jeb.082404
Mechanisms of muscle gene regulation in the electric organ of Sternopygus macrurus
Abstract
Animals perform a remarkable diversity of movements through the coordinated mechanical contraction of skeletal muscle. This capacity for a wide range of movements is due to the presence of muscle cells with a very plastic phenotype that display many different biochemical, physiological and morphological properties. What factors influence the maintenance and plasticity of differentiated muscle fibers is a fundamental question in muscle biology. We have exploited the remarkable potential of skeletal muscle cells of the gymnotiform electric fish Sternopygus macrurus to trans-differentiate into electrocytes, the non-contractile electrogenic cells of the electric organ (EO), to investigate the mechanisms that regulate the skeletal muscle phenotype. In S. macrurus, mature electrocytes possess a phenotype that is intermediate between muscle and non-muscle cells. How some genes coding for muscle-specific proteins are downregulated while others are maintained, and novel genes are upregulated, is an intriguing problem in the control of skeletal muscle and EO phenotype. To date, the intracellular and extracellular factors that generate and maintain distinct patterns of gene expression in muscle and EO have not been defined. Expression studies in S. macrurus have started to shed light on the role that transcriptional and post-transcriptional events play in regulating specific muscle protein systems and the muscle phenotype of the EO. In addition, these findings also represent an important step toward identifying mechanisms that affect the maintenance and plasticity of the muscle cell phenotype for the evolution of highly specialized non-contractile tissues.
Keywords: electrocyte; muscle regulatory factors; muscle-derived cells; post-transcriptional regulation.
Figures

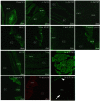
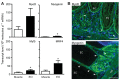
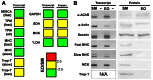
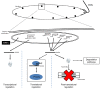
References
-
- Ai J., Zhang R., Gao X., Niu H.-F., Wang N., Xu Y., Li Y., Ma N., Sun L.-H., Pan Z.-W., et al. (2012). Overexpression of microRNA-1 impairs cardiac contractile function by damaging sarcomere assembly. Cardiovasc. Res. 95, 385-393 - PubMed
-
- Allen D. L., Sartorius C. A., Sycuro L. K., Leinwand L. A. (2001). Different pathways regulate expression of the skeletal myosin heavy chain genes. J. Biol. Chem. 276, 43524-43533 - PubMed
-
- Ambros V. (2004). The functions of animal microRNAs. Nature 431, 350-355 - PubMed
-
- Arnold H. H., Braun T. (1996). Targeted inactivation of myogenic factor genes reveals their role during mouse myogenesis: a review. Int. J. Dev. Biol. 40, 345-353 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

