Application of circuit simulation method for differential modeling of TIM-2 iron uptake and metabolism in mouse kidney cells
- PMID: 23761763
- PMCID: PMC3675319
- DOI: 10.3389/fphys.2013.00136
Application of circuit simulation method for differential modeling of TIM-2 iron uptake and metabolism in mouse kidney cells
Abstract
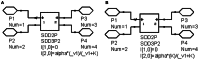
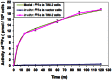
Circuit simulation is a powerful methodology to generate differential mathematical models. Due to its highly accurate modeling capability, circuit simulation can be used to investigate interactions between the parts and processes of a cellular system. Circuit simulation has become a core technology for the field of electrical engineering, but its application in biology has not yet been fully realized. As a case study for evaluating the more advanced features of a circuit simulation tool called Advanced Design System (ADS), we collected and modeled laboratory data for iron metabolism in mouse kidney cells for a H ferritin (HFt) receptor, T cell immunoglobulin and mucin domain-2 (TIM-2). The internal controlling parameters of TIM-2 associated iron metabolism were extracted and the ratios of iron movement among cellular compartments were quantified by ADS. The differential model processed by circuit simulation demonstrated a capability to identify variables and predict outcomes that could not be readily measured by in vitro experiments. For example, an initial rate of uptake of iron-loaded HFt (Fe-HFt) was 2.17 pmol per million cells. TIM-2 binding probability with Fe-HFt was 16.6%. An average of 8.5 min was required for the complex of TIM-2 and Fe-HFt to form an endosome. The endosome containing HFt lasted roughly 2 h. At the end of endocytosis, about 28% HFt remained intact and the rest was degraded. Iron released from degraded HFt was in the labile iron pool (LIP) and stimulated the generation of endogenous HFt for new storage. Both experimental data and the model showed that TIM-2 was not involved in the process of iron export. The extracted internal controlling parameters successfully captured the complexity of TIM-2 pathway and the use of circuit simulation-based modeling across a wider range of cellular systems is the next step for validating the significance and utility of this method.
Keywords: TIM-2; circuit simulator; export; ferritin; iron; model; storage; uptake.
Figures





Similar articles
-
Iron uptake mediated by binding of H-ferritin to the TIM-2 receptor in mouse cells.PLoS One. 2011;6(8):e23800. doi: 10.1371/journal.pone.0023800. Epub 2011 Aug 19. PLoS One. 2011. PMID: 21886823 Free PMC article.
-
Light-induced retinal degeneration correlates with changes in iron metabolism gene expression, ferritin level, and aging.Invest Ophthalmol Vis Sci. 2011 Mar 10;52(3):1261-74. doi: 10.1167/iovs.10-5705. Invest Ophthalmol Vis Sci. 2011. PMID: 20881284
-
TIM-2 is expressed on B cells and in liver and kidney and is a receptor for H-ferritin endocytosis.J Exp Med. 2005 Oct 3;202(7):955-65. doi: 10.1084/jem.20042433. J Exp Med. 2005. PMID: 16203866 Free PMC article.
-
The molecular mechanisms of the metabolism and transport of iron in normal and neoplastic cells.Biochim Biophys Acta. 1997 Mar 14;1331(1):1-40. doi: 10.1016/s0304-4157(96)00014-7. Biochim Biophys Acta. 1997. PMID: 9325434 Review.
-
The ferritins: molecular properties, iron storage function and cellular regulation.Biochim Biophys Acta. 1996 Jul 31;1275(3):161-203. doi: 10.1016/0005-2728(96)00022-9. Biochim Biophys Acta. 1996. PMID: 8695634 Review.
Cited by
-
Activated Oncogenic Pathway Modifies Iron Network in Breast Epithelial Cells: A Dynamic Modeling Perspective.PLoS Comput Biol. 2017 Feb 6;13(2):e1005352. doi: 10.1371/journal.pcbi.1005352. eCollection 2017 Feb. PLoS Comput Biol. 2017. PMID: 28166223 Free PMC article.
References
-
- Baynes R., Bukofzer G., Bothwell T., Bezwoda W., Macfarlane B. (1987). Transferrin receptors and transferrin iron uptake by cultured human blood monocytes. Eur. J. Cell Biol. 43, 372–376 - PubMed
-
- Han J., Day J. R., Thomson K., Connor J. R., Beard J. L. (2000). Iron deficiency alters H- and L-ferritin expression in rat brain. Cell Mol. Biol. (Noisy-le-grand) 46, 517–528 - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

