A digital compendium of genes mediating the reversible phosphorylation of proteins in fe-deficient Arabidopsis roots
- PMID: 23761801
- PMCID: PMC3669753
- DOI: 10.3389/fpls.2013.00173
A digital compendium of genes mediating the reversible phosphorylation of proteins in fe-deficient Arabidopsis roots
Abstract
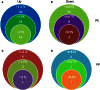
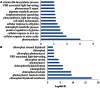
Post-translational modifications of proteins such as reversible phosphorylation provide an important but understudied regulatory network that controls important nodes in the adaptation of plants to environmental conditions. Iron (Fe) is an essential mineral nutrient for plants, but due to its low solubility often a limiting factor for optimal growth. To understand the role of protein phosphorylation in the regulation of cellular Fe homeostasis, we analyzed the expression of protein kinases (PKs) and phosphatases (PPs) in Arabidopsis roots by mining differentially expressed PK and PP genes. Transcriptome analysis using RNA-seq revealed that subsets of 203 PK and 39 PP genes were differentially expressed under Fe-deficient conditions. Functional modules of these PK and PP genes were further generated based on co-expression analysis using the MACCU toolbox on the basis of 300 publicly available root-related microarray data sets. Results revealed networks comprising 87 known or annotated PK and PP genes that could be subdivided into one large and several smaller highly co-expressed gene modules. The largest module was composed of 58 genes, most of which have been assigned to the leucine-rich repeat protein kinase superfamily and associated with the biological processes "hypotonic salinity response," "potassium ion import," and "cellular potassium ion homeostasis." The comprehensive transcriptional information on PK and PP genes in iron-deficient roots provided here sets the stage for follow-up experiments and contributes to our understanding of the post-translational regulation of Fe deficiency and potassium ion homeostasis.
Keywords: RNA-seq; co-expression; iron deficiency; potassium homeostasis; protein phosphorylation.
Figures






References
-
- Barak P., Chen Y. (1983). The effect of potassium fertilization on iron deficiency. Commun. Soil Sci. Plant Anal. 14, 945–950 10.1080/00103628309367422 - DOI
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

