DNA qualification workflow for next generation sequencing of histopathological samples
- PMID: 23762227
- PMCID: PMC3675123
- DOI: 10.1371/journal.pone.0062692
DNA qualification workflow for next generation sequencing of histopathological samples
Abstract
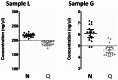
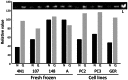
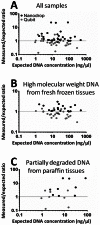
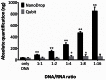
Histopathological samples are a treasure-trove of DNA for clinical research. However, the quality of DNA can vary depending on the source or extraction method applied. Thus a standardized and cost-effective workflow for the qualification of DNA preparations is essential to guarantee interlaboratory reproducible results. The qualification process consists of the quantification of double strand DNA (dsDNA) and the assessment of its suitability for downstream applications, such as high-throughput next-generation sequencing. We tested the two most frequently used instrumentations to define their role in this process: NanoDrop, based on UV spectroscopy, and Qubit 2.0, which uses fluorochromes specifically binding dsDNA. Quantitative PCR (qPCR) was used as the reference technique as it simultaneously assesses DNA concentration and suitability for PCR amplification. We used 17 genomic DNAs from 6 fresh-frozen (FF) tissues, 6 formalin-fixed paraffin-embedded (FFPE) tissues, 3 cell lines, and 2 commercial preparations. Intra- and inter-operator variability was negligible, and intra-methodology variability was minimal, while consistent inter-methodology divergences were observed. In fact, NanoDrop measured DNA concentrations higher than Qubit and its consistency with dsDNA quantification by qPCR was limited to high molecular weight DNA from FF samples and cell lines, where total DNA and dsDNA quantity virtually coincide. In partially degraded DNA from FFPE samples, only Qubit proved highly reproducible and consistent with qPCR measurements. Multiplex PCR amplifying 191 regions of 46 cancer-related genes was designated the downstream application, using 40 ng dsDNA from FFPE samples calculated by Qubit. All but one sample produced amplicon libraries suitable for next-generation sequencing. NanoDrop UV-spectrum verified contamination of the unsuccessful sample. In conclusion, as qPCR has high costs and is labor intensive, an alternative effective standard workflow for qualification of DNA preparations should include the sequential combination of NanoDrop and Qubit to assess the purity and quantity of dsDNA, respectively.
Conflict of interest statement
Figures

References
-
- Dong H, Wang S (2012) Exploring the cancer genome in the era of next-generation sequencing. Front Med 6: 48–55. - PubMed
-
- Ma QC, Ennis CA, Aparicio S (2012) Opening Pandora's Box – the new biology of driver mutations and clonal evolution in cancer as revealed by next generation sequencing. Curr Opin Genet Dev 22: 3–9. - PubMed
-
- Meyerson M, Gabriel S, Getz G (2010) Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11: 685–696. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

