Novel regulatory small RNAs in Streptococcus pyogenes
- PMID: 23762235
- PMCID: PMC3675131
- DOI: 10.1371/journal.pone.0064021
Novel regulatory small RNAs in Streptococcus pyogenes
Abstract
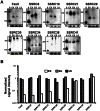
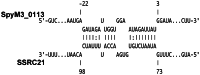
Streptococcus pyogenes (Group A Streptococcus or GAS) is a Gram-positive bacterial pathogen that has shown complex modes of regulation of its virulence factors to cause diverse diseases. Bacterial small RNAs are regarded as novel widespread regulators of gene expression in response to environmental signals. Recent studies have revealed that several small RNAs (sRNAs) have an important role in S. pyogenes physiology and pathogenesis by regulating gene expression at the translational level. To search for new sRNAs in S. pyogenes, we performed a genomewide analysis through computational prediction followed by experimental verification. To overcome the limitation of low accuracy in computational prediction, we employed a combination of three different computational algorithms (sRNAPredict, eQRNA and RNAz). A total of 45 candidates were chosen based on the computational analysis, and their transcription was analyzed by reverse-transcriptase PCR and Northern blot. Through this process, we discovered 7 putative novel trans-acting sRNAs. Their abundance varied between different growth phases, suggesting that their expression is influenced by environmental or internal signals. Further, to screen target mRNAs of an sRNA, we employed differential RNA sequencing analysis. This study provides a significant resource for future study of small RNAs and their roles in physiology and pathogenesis of S. pyogenes.
Conflict of interest statement
Figures



Similar articles
-
Identification of novel growth phase- and media-dependent small non-coding RNAs in Streptococcus pyogenes M49 using intergenic tiling arrays.BMC Genomics. 2012 Oct 13;13:550. doi: 10.1186/1471-2164-13-550. BMC Genomics. 2012. PMID: 23062031 Free PMC article.
-
Identification of streptococcal small RNAs that are putative targets of RNase III through bioinformatics analysis of RNA sequencing data.BMC Bioinformatics. 2017 Dec 28;18(Suppl 14):540. doi: 10.1186/s12859-017-1897-0. BMC Bioinformatics. 2017. PMID: 29297355 Free PMC article.
-
RNA sequencing uncovers antisense RNAs and novel small RNAs in Streptococcus pyogenes.RNA Biol. 2016;13(2):177-95. doi: 10.1080/15476286.2015.1110674. RNA Biol. 2016. PMID: 26580233 Free PMC article.
-
RNA-mediated regulation in Gram-positive pathogens: an overview punctuated with examples from the group A Streptococcus.Mol Microbiol. 2014 Oct;94(1):9-20. doi: 10.1111/mmi.12742. Epub 2014 Aug 21. Mol Microbiol. 2014. PMID: 25091277 Free PMC article. Review.
-
Genetic Regulation of Streptococci by Small RNAs.Curr Issues Mol Biol. 2019;32:39-86. doi: 10.21775/cimb.032.039. Epub 2019 Jun 5. Curr Issues Mol Biol. 2019. PMID: 31166169 Review.
Cited by
-
Dynamics of diversified A-to-I editing in Streptococcus pyogenes is governed by changes in mRNA stability.Nucleic Acids Res. 2024 Oct 14;52(18):11234-11253. doi: 10.1093/nar/gkae629. Nucleic Acids Res. 2024. PMID: 39087550 Free PMC article.
-
Regulatory RNAs in the Less Studied Streptococcal Species: From Nomenclature to Identification.Front Microbiol. 2016 Jul 26;7:1161. doi: 10.3389/fmicb.2016.01161. eCollection 2016. Front Microbiol. 2016. PMID: 27507970 Free PMC article. Review.
-
Dynamic Protein Phosphorylation in Streptococcus pyogenes during Growth, Stationary Phase, and Starvation.Microorganisms. 2024 Mar 20;12(3):621. doi: 10.3390/microorganisms12030621. Microorganisms. 2024. PMID: 38543672 Free PMC article.
-
Recent Research Advances in Small Regulatory RNAs in Streptococcus.Curr Microbiol. 2021 Jun;78(6):2231-2241. doi: 10.1007/s00284-021-02484-y. Epub 2021 May 7. Curr Microbiol. 2021. PMID: 33963446 Review.
-
The Second Messenger c-di-AMP Regulates Diverse Cellular Pathways Involved in Stress Response, Biofilm Formation, Cell Wall Homeostasis, SpeB Expression, and Virulence in Streptococcus pyogenes.Infect Immun. 2019 May 21;87(6):e00147-19. doi: 10.1128/IAI.00147-19. Print 2019 Jun. Infect Immun. 2019. PMID: 30936159 Free PMC article.
References
-
- Gripenland J, Netterling S, Loh E, Tiensuu T, Toledo-Arana A, et al. (2010) RNAs: regulators of bacterial virulence. Nat Rev Microbiol 8: 857–866. - PubMed
-
- Wassarman KM, Zhang A, Storz G (1999) Small RNAs in Escherichia coli . Trends Microbiol 7: 37–45. - PubMed
-
- Zhou Y, Xie J (2010) The roles of pathogen small RNAs. J Cell Physiol 226: 968–973. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

