Imputing amino acid polymorphisms in human leukocyte antigens
- PMID: 23762245
- PMCID: PMC3675122
- DOI: 10.1371/journal.pone.0064683
Imputing amino acid polymorphisms in human leukocyte antigens
Abstract
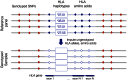
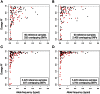
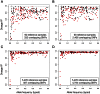
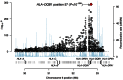
DNA sequence variation within human leukocyte antigen (HLA) genes mediate susceptibility to a wide range of human diseases. The complex genetic structure of the major histocompatibility complex (MHC) makes it difficult, however, to collect genotyping data in large cohorts. Long-range linkage disequilibrium between HLA loci and SNP markers across the major histocompatibility complex (MHC) region offers an alternative approach through imputation to interrogate HLA variation in existing GWAS data sets. Here we describe a computational strategy, SNP2HLA, to impute classical alleles and amino acid polymorphisms at class I (HLA-A, -B, -C) and class II (-DPA1, -DPB1, -DQA1, -DQB1, and -DRB1) loci. To characterize performance of SNP2HLA, we constructed two European ancestry reference panels, one based on data collected in HapMap-CEPH pedigrees (90 individuals) and another based on data collected by the Type 1 Diabetes Genetics Consortium (T1DGC, 5,225 individuals). We imputed HLA alleles in an independent data set from the British 1958 Birth Cohort (N = 918) with gold standard four-digit HLA types and SNPs genotyped using the Affymetrix GeneChip 500 K and Illumina Immunochip microarrays. We demonstrate that the sample size of the reference panel, rather than SNP density of the genotyping platform, is critical to achieve high imputation accuracy. Using the larger T1DGC reference panel, the average accuracy at four-digit resolution is 94.7% using the low-density Affymetrix GeneChip 500 K, and 96.7% using the high-density Illumina Immunochip. For amino acid polymorphisms within HLA genes, we achieve 98.6% and 99.3% accuracy using the Affymetrix GeneChip 500 K and Illumina Immunochip, respectively. Finally, we demonstrate how imputation and association testing at amino acid resolution can facilitate fine-mapping of primary MHC association signals, giving a specific example from type 1 diabetes.
Conflict of interest statement
Figures
References
-
- Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, et al. (2004) Gene map of the extended human MHC. Nature reviews Genetics 5: 889–899. - PubMed
-
- Carrington M, O’Brien SJ (2003) The influence of HLA genotype on AIDS. Annu Rev Med 54: 535–551. - PubMed
-
- Morishima S, Ogawa S, Matsubara A, Kawase T, Nannya Y, et al. (2010) Impact of highly conserved HLA haplotype on acute graft-versus-host disease. Blood 115: 4664–4670. - PubMed
-
- Bharadwaj M, Illing P, Theodossis A, Purcell AW, Rossjohn J, et al. (2012) Drug hypersensitivity and human leukocyte antigens of the major histocompatibility complex. Annu Rev Pharmacol Toxicol 52: 401–431. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

