Effects of HCV on basal and tat-induced HIV LTR activation
- PMID: 23762271
- PMCID: PMC3677892
- DOI: 10.1371/journal.pone.0064956
Effects of HCV on basal and tat-induced HIV LTR activation
Abstract
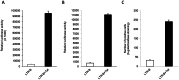
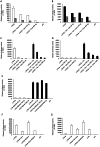
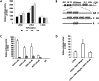
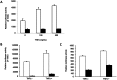
Hepatitis C virus (HCV) co-infection occurs in ∼30-40% of the HIV-infected population in the US. While a significant body of research suggests an adverse effect of HIV on HCV replication and disease progression, the impact of HCV on HIV infection has not been well studied. Increasing data suggest that hepatocytes and other liver cell populations can serve as reservoirs for HIV replication. Therefore, to gain insight into the impact of HCV on HIV, the effects of the HCV Core protein and infectious hepatitis C virions were evaluated on basal and Tat-induced activation of the HIV long terminal repeat (LTR) in hepatocytes. The HIV LTR was highly induced by the HIV transactivator protein Tat in hepatocytes. Activation varied according to the number of NF-kB binding sites present in the LTRs from different HIV subtypes. Involvement of the NF-kB binding pathway in LTR activation was demonstrated using an NF-kB inhibitor and deletion of the NF-kB binding sites. TNFα, a pro-inflammatory cytokine that plays an important role in HIV pathogenesis, also induced LTR activity in hepatocytes. However, HIV LTR activity was suppressed in hepatocytes in the presence of HCV Core protein, and the suppressive effect persisted in the presence of TNFα. In contrast, infectious hepatitis C virions upregulated HIV LTR activation and gene transcription. Core-mediated suppression remained unaltered in the presence of HCV NS3/4A protein, suggesting the involvement of other viral/cellular factors. These findings have significant clinical implications as they imply that HCV could accelerate HIV disease progression in HIV/HCV co-infected patients. Such analyses are important to elucidate the mechanisms by which these viruses interact and could facilitate the development of more effective therapies to treat HIV/HCV co-infection.
Conflict of interest statement
Figures


References
-
- Lavanchy D (2009) The global burden of hepatitis C. Liver International. 29: 74–81. - PubMed
-
- Alter M, Druszon-Moran D, Nainan O, McQuillan G, Gao F, et al. (1999) The prevalence of hepatitis C virus infection in the United States, 1988 through 1994. New England Journal of Medicine 341: 556–562. - PubMed
-
- Sherman K, Rouster S, Chung R, Rajicic N (2002) Hepatitis C virus prevalence among patients infected with human immunodeficiency virus: a cross-sectional analysis of the US adult AIDS clinical trials groups. Clinical Infectious Diseases 34: 831–837. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

