MiR-7 triggers cell cycle arrest at the G1/S transition by targeting multiple genes including Skp2 and Psme3
- PMID: 23762407
- PMCID: PMC3675065
- DOI: 10.1371/journal.pone.0065671
MiR-7 triggers cell cycle arrest at the G1/S transition by targeting multiple genes including Skp2 and Psme3
Abstract
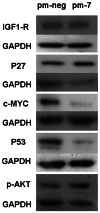
MiR-7 acts as a tumour suppressor in many cancers and abrogates proliferation of CHO cells in culture. In this study we demonstrate that miR-7 targets key regulators of the G1 to S phase transition, including Skp2 and Psme3, to promote increased levels of p27(KIP) and temporary growth arrest of CHO cells in the G1 phase. Simultaneously, the down-regulation of DNA repair-specific proteins via miR-7 including Rad54L, and pro-apoptotic regulators such as p53, combined with the up-regulation of anti-apoptotic factors like p-Akt, promoted cell survival while arrested in G1. Thus miR-7 can co-ordinate the levels of multiple genes and proteins to influence G1 to S phase transition and the apoptotic response in order to maintain cellular homeostasis. This work provides further mechanistic insight into the role of miR-7 as a regulator of cell growth in times of cellular stress.
Conflict of interest statement
Figures
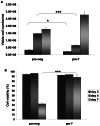
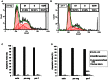




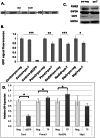
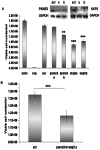
References
-
- Ichimura A, Ruike Y, Terasawa K, Tsujimoto G (2011) miRNAs and regulation of cell signaling. FEBS J 278: 1610–8. - PubMed
-
- Cannell IG, Bushell M (2010) Regulation of myc by miR-34c: A mechanism to prevent genomic instability? Cell Cycle 9: 2726–30. - PubMed
-
- Lewis BP, Burge CB, Bartel DP (2008) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

