nfxB as a novel target for analysis of mutation spectra in Pseudomonas aeruginosa
- PMID: 23762483
- PMCID: PMC3676378
- DOI: 10.1371/journal.pone.0066236
nfxB as a novel target for analysis of mutation spectra in Pseudomonas aeruginosa
Abstract
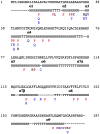
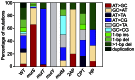
nfxB encodes a negative regulator of the mexCD-oprJ genes for drug efflux in the opportunistic pathogen Pseudomonas aeruginosa. Inactivating mutations in this transcriptional regulator constitute one of the main mechanisms of resistance to ciprofloxacin (Cip(r)). In this work, we evaluated the use of nfxB/Cip(r) as a new test system to study mutation spectra in P. aeruginosa. The analysis of 240 mutations in nfxB occurring spontaneously in the wild-type and mutator backgrounds or induced by mutagens showed that nfxB/Cip(r) offers several advantages compared with other mutation detection systems. Identification of nfxB mutations was easy since the entire open reading frame and its promoter region were sequenced from the chromosome using a single primer. Mutations detected in nfxB included all transitions and transversions, 1-bp deletions and insertions, >1-bp deletions and duplications. The broad mutation spectrum observed in nfxB relies on the selection of loss-of-function changes, as we confirmed by generating a structural model of the NfxB repressor and evaluating the significance of each detected mutation. The mutation spectra characterized in the mutS, mutT, mutY and mutM mutator backgrounds or induced by the mutagenic agents 2-aminopurine, cisplatin and hydrogen peroxide were in agreement with their predicted mutational specificities. Additionally, this system allowed the analysis of sequence context effects since point mutations occurred at 85 different sites distributed over the entire nfxB. Significant hotspots and preferred sequence contexts were observed for spontaneous and mutagen-induced mutation spectra. Finally, we demonstrated the utility of a luminescence-based reporter for identification of nfxB mutants previous to sequencing analysis. Thus, the nfxB/Cip(r) system in combination with the luminescent reporter may be a valuable tool for studying mutational processes in Pseudomonas spp. wherein the genes encoding the NfxB repressor and the associated efflux pump are conserved.
Conflict of interest statement
Figures



Similar articles
-
Real-Time Monitoring of nfxB Mutant Occurrence and Dynamics in Pseudomonas aeruginosa Biofilm Exposed to Subinhibitory Concentrations of Ciprofloxacin.Antimicrob Agents Chemother. 2017 Feb 23;61(3):e02292-16. doi: 10.1128/AAC.02292-16. Print 2017 Mar. Antimicrob Agents Chemother. 2017. PMID: 27993856 Free PMC article.
-
Functional characterization of the NfxB repressor of the mexCD-oprJ multidrug efflux operon of Pseudomonas aeruginosa.Microbiology (Reading). 2013 Oct;159(Pt 10):2058-2073. doi: 10.1099/mic.0.069286-0. Epub 2013 Aug 7. Microbiology (Reading). 2013. PMID: 23924707
-
Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa.Mol Microbiol. 1996 Aug;21(4):713-24. doi: 10.1046/j.1365-2958.1996.281397.x. Mol Microbiol. 1996. PMID: 8878035
-
Parallel evolution and local differentiation in quinolone resistance in Pseudomonas aeruginosa.Microbiology (Reading). 2011 Apr;157(Pt 4):937-944. doi: 10.1099/mic.0.046870-0. Epub 2011 Feb 3. Microbiology (Reading). 2011. PMID: 21292748 Review.
-
Mechanisms of ciprofloxacin resistance in Pseudomonas aeruginosa: new approaches to an old problem.J Med Microbiol. 2019 Jan;68(1):1-10. doi: 10.1099/jmm.0.000873. J Med Microbiol. 2019. PMID: 30605076 Review.
Cited by
-
The methylation-independent mismatch repair machinery in Pseudomonas aeruginosa.Microbiology (Reading). 2021 Dec;167(12):001120. doi: 10.1099/mic.0.001120. Microbiology (Reading). 2021. PMID: 34882086 Free PMC article. Review.
-
A role for the stringent response in ciprofloxacin resistance in Pseudomonas aeruginosa.Sci Rep. 2024 Apr 13;14(1):8598. doi: 10.1038/s41598-024-59188-z. Sci Rep. 2024. PMID: 38615146 Free PMC article.
-
Shared and Unique Evolutionary Trajectories to Ciprofloxacin Resistance in Gram-Negative Bacterial Pathogens.mBio. 2021 Jun 29;12(3):e0098721. doi: 10.1128/mBio.00987-21. Epub 2021 Jun 22. mBio. 2021. PMID: 34154405 Free PMC article.
-
The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria.Clin Microbiol Rev. 2015 Apr;28(2):337-418. doi: 10.1128/CMR.00117-14. Clin Microbiol Rev. 2015. PMID: 25788514 Free PMC article. Review.
-
In Vitro Activity of Cefepime-Taniborbactam and Comparators against Clinical Isolates of Gram-Negative Bacilli from 2018 to 2020: Results from the Global Evaluation of Antimicrobial Resistance via Surveillance (GEARS) Program.Antimicrob Agents Chemother. 2023 Jan 24;67(1):e0128122. doi: 10.1128/aac.01281-22. Epub 2022 Dec 21. Antimicrob Agents Chemother. 2023. PMID: 36541767 Free PMC article.
References
-
- Maki H (2002) Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu Rev Genet 36: 279–303. - PubMed
-
- Miller JH (1996) Spontaneous mutators in bacteria: insights into pathways of mutagenesis and repair. Annu Rev Microbiol 50: 625–643. - PubMed
-
- Peix A, Ramirez-Bahena MH, Velazquez E (2009) Historical evolution and current status of the taxonomy of genus Pseudomonas . Infect Genet Evol 9: 1132–1147. - PubMed
-
- Silby MW, Winstanley C, Godfrey SA, Levy SB, Jackson RW (2011) Pseudomonas genomes: diverse and adaptable. FEMS Microbiol Rev 35: 652–680. - PubMed
-
- Driscoll JA, Brody SL, Kollef MH (2007) The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67: 351–368. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

