Low osteogenic differentiation potential of placenta-derived mesenchymal stromal cells correlates with low expression of the transcription factors Runx2 and Twist2
- PMID: 23763516
- PMCID: PMC3804084
- DOI: 10.1089/scd.2012.0693
Low osteogenic differentiation potential of placenta-derived mesenchymal stromal cells correlates with low expression of the transcription factors Runx2 and Twist2
Abstract
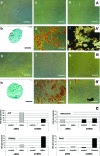
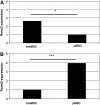
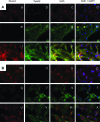
Recent studies indicated that mesenchymal stromal cells from bone marrow (bmMSC) differ in their osteogenic differentiation capacity compared to MSC from term placenta (pMSC). We extended these studies and investigated the expression of factors involved in regulation of bone metabolism in both cell types. To this end, MSC were expanded in vitro and characterized. The total transcriptome was investigated by microarrays, and for selected genes, the differences in gene expression were explored by quantitative reverse transcriptase-polymerase chain reaction, immunocytochemistry, and flow cytometry. We report that bmMSC and pMSC share expression of typical lineage surface markers, including CD73, CD90, CD105, and lack of CD14, CD34, and CD45. However, according to transcriptome analyses, they differ significantly in their expression of more than 590 genes. Factors involved in bone metabolism, including alkaline phosphatase (P<0.05), osteoglycin (P<0.05), osteomodulin (P<0.05), runt-related transcription factor 2 (Runx2) (P<0.04), and WISP2 (P<0.05), were expressed at significantly lower levels in pMSC, but twist-related protein 2 (Twist2) (P<0.0002) was expressed at significantly higher levels. The osteogenic differentiation capacity of pMSC was very low. The adipogenic differentiation was somewhat more prominent in bmMSC, while the chondrogenic differentiation seemed not to differ between bmMSC and pMSC, as determined by histochemical staining. However, expression and induction of peroxisome proliferator-activated receptor gamma-2 (PPARγ2) and Sox9, factors involved in early adipogenesis and chondrogenesis, respectively, were higher in bmMSC. We conclude that despite many similarities between bmMSC and pMSC, when expanded under identical conditions, they vary considerably with respect to their in vitro differentiation potential. For regenerative purposes, the choice of MSC may therefore influence the outcome of a treatment considerably.
Figures




References
-
- Dominici M. Le Blanc K. Mueller I. Slaper-Cortenbach I. Marini F. Krause D. Deans R. Keating A. Prockop D. Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. - PubMed
-
- Parolini O. Alviano F. Bagnara GP. Bilic G. Buhring HJ. Evangelista M. Hennerbichler S. Liu B. Magatti M, et al. Concise review: isolation and characterization of cells from human term placenta: outcome of the first International Workshop on Placenta Derived Stem Cells. Stem Cells. 2008;26:300–311. - PubMed
-
- Chamberlain G. Fox J. Ashton B. Middleton J. Concise review: mesenchymal stem cells: their phenotype, differentiation capacity, immunological features, and potential for homing. Stem Cells. 2007;25:2739–2749. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

