PDEF promotes luminal differentiation and acts as a survival factor for ER-positive breast cancer cells
- PMID: 23764000
- PMCID: PMC3711136
- DOI: 10.1016/j.ccr.2013.04.026
PDEF promotes luminal differentiation and acts as a survival factor for ER-positive breast cancer cells
Abstract
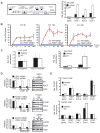
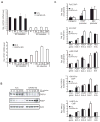
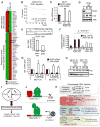
Breast cancer is a heterogeneous disease and can be classified based on gene expression profiles that reflect distinct epithelial subtypes. We identify prostate-derived ETS factor (PDEF) as a mediator of mammary luminal epithelial lineage-specific gene expression and as a factor required for tumorigenesis in a subset of breast cancers. PDEF levels strongly correlate with estrogen receptor (ER)-positive luminal breast cancer, and PDEF transcription is inversely regulated by ER and GATA3. Furthermore, PDEF is essential for luminal breast cancer cell survival and is required in models of endocrine resistance. These results offer insights into the function of this ETS factor that are clinically relevant and may be of therapeutic value for patients with breast cancer treated with endocrine therapy.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures





References
-
- Allinen M, Beroukhim R, Cai L, Brennan C, Lahti-Domenici J, Huang H, Porter D, Hu M, Chin L, Richardson A, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer cell. 2004;6:17–32. - PubMed
-
- Ariazi EA, Cunliffe HE, Lewis-Wambi JS, Slifker MJ, Willis AL, Ramos P, Tapia C, Kim HR, Yerrum S, Sharma CG, et al. Estrogen induces apoptosis in estrogen deprivation-resistant breast cancer through stress responses as identified by global gene expression across time. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18879–18886. - PMC - PubMed
-
- Asselin-Labat ML, Sutherland KD, Barker H, Thomas R, Shackleton M, Forrest NC, Hartley L, Robb L, Grosveld FG, van der Wees J, et al. Gata-3 is an essential regulator of mammary-gland morphogenesis and luminal-cell differentiation. Nature cell biology. 2007;9:201–209. - PubMed
-
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nature genetics. 2006;38:1289–1297. - PubMed
-
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer cell. 2006;10:529–541. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

