Methylation levels of LINE-1 in primary lesion and matched metastatic lesions of colorectal cancer
- PMID: 23764749
- PMCID: PMC3721399
- DOI: 10.1038/bjc.2013.289
Methylation levels of LINE-1 in primary lesion and matched metastatic lesions of colorectal cancer
Abstract
Background: LINE-1 methylation level is a surrogate marker of global DNA methylation. LINE-1 methylation in primary colorectal cancers (CRCs) is highly variable and strongly associated with a poor prognosis. However, no study has examined LINE-1 methylation levels of metastatic CRCs in relation to prognosis or assessed the heterogeneity of LINE-1 methylation level within the primary CRCs.
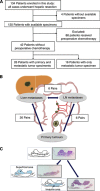
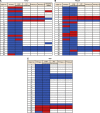
Methods: Pyrosequencing was used to quantify LINE-1 methylation level in 42 liver metastases, 26 matched primary tumours, and 6 matched lymph node (LN) metastases. KRAS, BRAF, and PIK3CA mutation status and microsatellite instability (MSI) status were also examined.
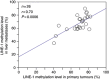
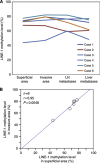
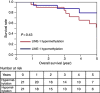
Results: The distribution of LINE-1 methylation level in liver metastases was as follows: mean, 67.3; range, 37.1-90.1. Primary tumours showed LINE-1 methylation levels similar to those of matched liver and LN metastases. The difference in LINE-1 methylation level between superficial areas and invasive front areas was within 7.0 in all six cases evaluated. Prognostic impact of LINE-1 hypomethylation in liver metastases on overall survival was not observed. The concordance rate was 94% for KRAS, 100% for BRAF, 88% for PIK3CA, and 97% for MSI.
Conclusion: Alteration of LINE-1 methylation level may occur in early CRC tumorigenesis, and the LINE-1 methylation level is relatively stable during CRC progression.
Figures
Similar articles
-
Tumor LINE-1 methylation level and microsatellite instability in relation to colorectal cancer prognosis.J Natl Cancer Inst. 2014 Sep 4;106(9):dju195. doi: 10.1093/jnci/dju195. Print 2014 Sep. J Natl Cancer Inst. 2014. PMID: 25190725 Free PMC article.
-
Clinical outcomes of patients with microsatellite-unstable colorectal carcinomas depend on L1 methylation level.Ann Surg Oncol. 2012 Oct;19(11):3441-8. doi: 10.1245/s10434-012-2410-7. Epub 2012 May 23. Ann Surg Oncol. 2012. PMID: 22618722
-
LINE-1 methylation shows little intra-patient heterogeneity in primary and synchronous metastatic colorectal cancer.BMC Cancer. 2012 Dec 5;12:574. doi: 10.1186/1471-2407-12-574. BMC Cancer. 2012. PMID: 23216958 Free PMC article.
-
High concordance rate of KRAS/BRAF mutations and MSI-H between primary colorectal cancer and corresponding metastases.Oncol Rep. 2017 Feb;37(2):785-792. doi: 10.3892/or.2016.5323. Epub 2016 Dec 15. Oncol Rep. 2017. PMID: 28000889
-
LINE-1 methylation level and prognosis in pancreas cancer: pyrosequencing technology and literature review.Surg Today. 2017 Dec;47(12):1450-1459. doi: 10.1007/s00595-017-1539-1. Epub 2017 May 23. Surg Today. 2017. PMID: 28536860 Review.
Cited by
-
DNA Methylation and Hydroxymethylation in Primary Colon Cancer and Synchronous Hepatic Metastasis.Front Genet. 2018 Jan 9;8:229. doi: 10.3389/fgene.2017.00229. eCollection 2017. Front Genet. 2018. PMID: 29375619 Free PMC article.
-
Technical advances in global DNA methylation analysis in human cancers.J Biol Eng. 2017 Mar 1;11:10. doi: 10.1186/s13036-017-0052-9. eCollection 2017. J Biol Eng. 2017. PMID: 28261325 Free PMC article. Review.
-
Decoupling of DNA methylation and activity of intergenic LINE-1 promoters in colorectal cancer.Epigenetics. 2017 Jun 3;12(6):465-475. doi: 10.1080/15592294.2017.1300729. Epub 2017 Mar 16. Epigenetics. 2017. PMID: 28300471 Free PMC article.
-
TET family proteins and 5-hydroxymethylcytosine in esophageal squamous cell carcinoma.Oncotarget. 2015 Sep 15;6(27):23372-82. doi: 10.18632/oncotarget.4281. Oncotarget. 2015. PMID: 26093090 Free PMC article.
-
Biomarker concordance between primary colorectal cancer and its metastases.EBioMedicine. 2019 Feb;40:363-374. doi: 10.1016/j.ebiom.2019.01.050. Epub 2019 Feb 4. EBioMedicine. 2019. PMID: 30733075 Free PMC article.
References
-
- Albanese I, Scibetta AG, Migliavacca M, Russo A, Bazan V, Tomasino RM, Colomba P, Tagliavia M, La Farina M. Heterogeneity within and between primary colorectal carcinomas and matched metastases as revealed by analysis of Ki-ras and p53 mutations. Biochem Biophys Res Commun. 2004;325:784–791. - PubMed
-
- Allegra CJ, Jessup JM, Somerfield MR, Hamilton SR, Hammond EH, Hayes DF, McAllister PK, Morton RF, Schilsky RL. American Society of Clinical Oncology provisional clinical opinion: testing for KRAS gene mutations in patients with metastatic colorectal carcinoma to predict response to anti-epidermal growth factor receptor monoclonal antibody therapy. J Clin Oncol. 2009;27:2091–2096. - PubMed
-
- Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, Juan T, Sikorski R, Suggs S, Radinsky R, Patterson SD, Chang DD. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. - PubMed
-
- Benvenuti S, Sartore-Bianchi A, Di Nicolantonio F, Zanon C, Moroni M, Veronese S, Siena S, Bardelli A. Oncogenic activation of the RAS/RAF signaling pathway impairs the response of metastatic colorectal cancers to anti-epidermal growth factor receptor antibody therapies. Cancer Res. 2007;67:2643–2648. - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

