Chromatin and epigenetic features of long-range gene regulation
- PMID: 23766291
- PMCID: PMC3753629
- DOI: 10.1093/nar/gkt499
Chromatin and epigenetic features of long-range gene regulation
Abstract
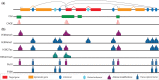
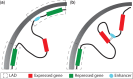
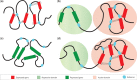
The precise regulation of gene transcription during metazoan development is controlled by a complex system of interactions between transcription factors, histone modifications and modifying enzymes and chromatin conformation. Developments in chromosome conformation capture technologies have revealed that interactions between regions of chromatin are pervasive and highly cell-type specific. The movement of enhancers and promoters in and out of higher-order chromatin structures within the nucleus are associated with changes in expression and histone modifications. However, the factors responsible for mediating these changes and determining enhancer:promoter specificity are still not completely known. In this review, we summarize what is known about the patterns of epigenetic and chromatin features characteristic of elements involved in long-range interactions. In addition, we review the insights into both local and global patterns of chromatin interactions that have been revealed by the latest experimental and computational methods.
Figures
References
-
- Pearson JC, Lemons D, McGinnis W. Modulating Hox gene functions during animal body patterning. Nat. Rev. Genet. 2005;6:893–904. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

