Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome
- PMID: 23767915
- PMCID: PMC3707022
- DOI: 10.1186/gb-2013-14-6-r59
Hyperosmotic priming of Arabidopsis seedlings establishes a long-term somatic memory accompanied by specific changes of the epigenome
Abstract
Background: In arid and semi-arid environments, drought and soil salinity usually occur at the beginning and end of a plant's life cycle, offering a natural opportunity for the priming of young plants to enhance stress tolerance in mature plants. Chromatin marks, such as histone modifications, provide a potential molecular mechanism for priming plants to environmental stresses, but whether transient exposure of seedlings to hyperosmotic stress leads to chromatin changes that are maintained throughout vegetative growth remains unclear.
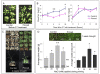
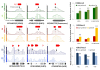
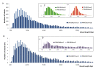
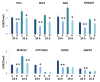
Results: We have established an effective protocol for hyperosmotic priming in the model plant Arabidopsis, which includes a transient mild salt treatment of seedlings followed by an extensive period of growth in control conditions. Primed plants are identical to non-primed plants in growth and development, yet they display reduced salt uptake and enhanced drought tolerance after a second stress exposure. ChIP-seq analysis of four histone modifications revealed that the priming treatment altered the epigenomic landscape; the changes were small but they were specific for the treated tissue, varied in number and direction depending on the modification, and preferentially targeted transcription factors. Notably, priming leads to shortening and fractionation of H3K27me3 islands. This effect fades over time, but is still apparent after a ten day growth period in control conditions. Several genes with priming-induced differences in H3K27me3 showed altered transcriptional responsiveness to the second stress treatment.
Conclusion: Experience of transient hyperosmotic stress by young plants is stored in a long-term somatic memory comprising differences of chromatin status, transcriptional responsiveness and whole plant physiology.
Figures






References
-
- Ventura L, Dona M, Macovei A, Carbonera D, Buttafava A, Mondoni A, Rossi G, Balestrazzi A. Understanding the molecular pathways associated with seed vigor. Plant Physiol Biochem. 2012;60:196–206. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

