Molecular profiling of human mammary gland links breast cancer risk to a p27(+) cell population with progenitor characteristics
- PMID: 23770079
- PMCID: PMC3703476
- DOI: 10.1016/j.stem.2013.05.004
Molecular profiling of human mammary gland links breast cancer risk to a p27(+) cell population with progenitor characteristics
Abstract
Early full-term pregnancy is one of the most effective natural protections against breast cancer. To investigate this effect, we have characterized the global gene expression and epigenetic profiles of multiple cell types from normal breast tissue of nulliparous and parous women and carriers of BRCA1 or BRCA2 mutations. We found significant differences in CD44(+) progenitor cells, where the levels of many stem cell-related genes and pathways, including the cell-cycle regulator p27, are lower in parous women without BRCA1/BRCA2 mutations. We also noted a significant reduction in the frequency of CD44(+)p27(+) cells in parous women and showed, using explant cultures, that parity-related signaling pathways play a role in regulating the number of p27(+) cells and their proliferation. Our results suggest that pathways controlling p27(+) mammary epithelial cells and the numbers of these cells relate to breast cancer risk and can be explored for cancer risk assessment and prevention.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



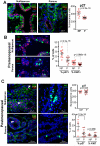
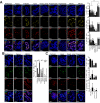

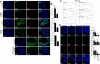
Comment in
-
Pregnancy offers new insights into mechanisms of breast cancer risk and resistance.Breast Cancer Res. 2013;15(5):312. doi: 10.1186/bcr3482. Breast Cancer Res. 2013. PMID: 24060354 Free PMC article.
References
-
- Asselin-Labat ML, Vaillant F, Sheridan JM, Pal B, Wu D, Simpson ER, Yasuda H, Smyth GK, Martin TJ, Lindeman GJ, et al. Control of mammary stem cell function by steroid hormone signalling. Nature. 2010;465:798–802. - PubMed
-
- Asztalos S, Gann PH, Hayes MK, Nonn L, Beam CA, Dai Y, Wiley EL, Tonetti DA. Gene expression patterns in the human breast after pregnancy. Cancer Prev Res (Phila) 2010;3:301–311. - PubMed
-
- Blakely CM, Stoddard AJ, Belka GK, Dugan KD, Notarfrancesco KL, Moody SE, D'Cruz CM, Chodosh LA. Hormone-induced protection against mammary tumorigenesis is conserved in multiple rat strains and identifies a core gene expression signature induced by pregnancy. Cancer Res. 2006;66:6421–6431. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

